
The ribosome is a complex molecular machine, found within all living cells, that serves as the site of biological protein synthesis (translation). Ribosomes link amino acids together in the order specified by messenger RNA (mRNA) molecules. Ribosomes consist of two major components: the small ribosomal subunits, which read the RNA, and the large subunits, which join amino acids to form a polypeptide chain. Each subunit comprises one or more ribosomal RNA (rRNA) molecules and a variety of ribosomal proteins. The ribosomes and associated molecules are also known as the translational apparatus.

In molecular biology and genetics, translation is the process in which ribosomes in the cytoplasm or ER synthesize proteins after the process of transcription of DNA to RNA in the cell's nucleus. The entire process is called gene expression.
Prokaryotic translation is the process by which messenger RNA is translated into proteins in prokaryotes.
Eukaryotic translation is the biological process by which messenger RNA is translated into proteins in eukaryotes. It consists of four phases: initiation, elongation, termination, and recycling.
A release factor is a protein that allows for the termination of translation by recognizing the termination codon or stop codon in an mRNA sequence.
Eukaryotic initiation factors (eIFs) are proteins or protein complexes involved in the initiation phase of eukaryotic translation. These proteins help stabilize the formation of ribosomal preinitiation complexes around the start codon and are an important input for post-transcription gene regulation. Several initiation factors form a complex with the small 40S ribosomal subunit and Met-tRNAiMet called the 43S preinitiation complex. Additional factors of the eIF4F complex recruit the 43S PIC to the five-prime cap structure of the mRNA, from which the 43S particle scans 5'-->3' along the mRNA to reach an AUG start codon. Recognition of the start codon by the Met-tRNAiMet promotes gated phosphate and eIF1 release to form the 48S preinitiation complex, followed by large 60S ribosomal subunit recruitment to form the 80S ribosome. There exist many more eukaryotic initiation factors than prokaryotic initiation factors, reflecting the greater biological complexity of eukaryotic translation. There are at least twelve eukaryotic initiation factors, composed of many more polypeptides, and these are described below.

Eukaryotic translation termination factor 1 (eRF1), also known asTB3-1, is a protein that in humans is encoded by the ETF1 gene.

DEAD box proteins are involved in an assortment of metabolic processes that typically involve RNAs, but in some cases also other nucleic acids. They are highly conserved in nine motifs and can be found in most prokaryotes and eukaryotes, but not all. Many organisms, including humans, contain DEAD-box helicases, which are involved in RNA metabolism.

Eukaryotic translation initiation factor 2 subunit 2 (eIF2β) is a protein that in humans is encoded by the EIF2S2 gene.

Eukaryotic peptide chain release factor GTP-binding subunit ERF3A is an enzyme that in humans is encoded by the GSPT1 gene.

Eukaryotic translation initiation factor 3 subunit D (eIF3d) is a protein that in humans is encoded by the EIF3D gene.

EF-G is a prokaryotic elongation factor involved in protein translation. As a GTPase, EF-G catalyzes the movement (translocation) of transfer RNA (tRNA) and messenger RNA (mRNA) through the ribosome.

Eukaryotic peptide chain release factor GTP-binding subunit ERF3B is an enzyme that in humans is encoded by the GSPT2 gene.
Eukaryotic translation initiation factor 4 G (eIF4G) is a protein involved in eukaryotic translation initiation and is a component of the eIF4F cap-binding complex. Orthologs of eIF4G have been studied in multiple species, including humans, yeast, and wheat. However, eIF4G is exclusively found in domain Eukarya, and not in domains Bacteria or Archaea, which do not have capped mRNA. As such, eIF4G structure and function may vary between species, although the human eIF4G 1 has been the focus of extensive studies.
Eukaryotic Initiation Factor 2 (eIF2) is a eukaryotic initiation factor. It is required for most forms of eukaryotic translation initiation. eIF2 mediates the binding of tRNAiMet to the ribosome in a GTP-dependent manner. eIF2 is a heterotrimer consisting of an alpha, a beta, and a gamma subunit.
Translational regulation refers to the control of the levels of protein synthesized from its mRNA. This regulation is vastly important to the cellular response to stressors, growth cues, and differentiation. In comparison to transcriptional regulation, it results in much more immediate cellular adjustment through direct regulation of protein concentration. The corresponding mechanisms are primarily targeted on the control of ribosome recruitment on the initiation codon, but can also involve modulation of peptide elongation, termination of protein synthesis, or ribosome biogenesis. While these general concepts are widely conserved, some of the finer details in this sort of regulation have been proven to differ between prokaryotic and eukaryotic organisms.
NatA acetyltransferase(Nα acetyltransferase), is an enzyme that serves to catalyze the addition of acetyl groups to various proteins emerging from the ribosome. Upon translation, the NatA binds to the ribosome and then "stretches" to the front end of the forming, or nascent, polypeptide, where it adds this acetyl group. This acetyl group is added to the front end, or N-terminus of the new protein.
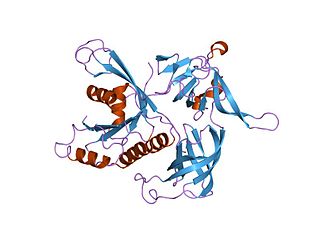
In molecular biology, the GTP-binding elongation factor family, EF-Tu/EF-1A subfamily is a family of elongation factors, which includes the eukaryotic eEF-1 and the prokaryotic EF-Tu.
Eukaryotic initiation factor 4F (eIF4F) is a heterotrimeric protein complex that binds the 5' cap of messenger RNAs (mRNAs) to promote eukaryotic translation initiation. The eIF4F complex is composed of three non-identical subunits: the DEAD-box RNA helicase eIF4A, the cap-binding protein eIF4E, and the large "scaffold" protein eIF4G. The mammalian eIF4F complex was first described in 1983, and has been a major area of study into the molecular mechanisms of cap-dependent translation initiation ever since.











