Related Research Articles

In cell biology, mitosis is a part of the cell cycle in which replicated chromosomes are separated into two new nuclei. Cell division gives rise to genetically identical cells in which the total number of chromosomes is maintained. Therefore, the mitosis is also known as equational division. In general, mitosis is preceded by the S stage of interphase and is often followed by telophase and cytokinesis; which divides the cytoplasm, organelles and cell membrane of one cell into two new cells containing roughly equal shares of these cellular components. The different stages of Mitosis altogether define the mitotic (M) phase of an animal cell cycle—the division of the mother cell into two daughter cells genetically identical to each other.
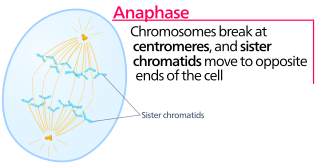
Anaphase, is the stage of mitosis after the process of metaphase, when replicated chromosomes are split and the newly-copied chromosomes are moved to opposite poles of the cell. Chromosomes also reach their overall maximum condensation in late anaphase, to help chromosome segregation and the re-formation of the nucleus.

In cell biology, the spindle apparatus refers to the cytoskeletal structure of eukaryotic cells that forms during cell division to separate sister chromatids between daughter cells. It is referred to as the mitotic spindle during mitosis, a process that produces genetically identical daughter cells, or the meiotic spindle during meiosis, a process that produces gametes with half the number of chromosomes of the parent cell.
Cyclin is a family of proteins that controls the progression of a cell through the cell cycle by activating cyclin-dependent kinase (CDK) enzymes or group of enzymes required for synthesis of cell cycle.

The spindle checkpoint, also known as the metaphase-to-anaphase transition, the spindle assembly checkpoint (SAC), or the mitotic checkpoint, is a cell cycle checkpoint during mitosis or meiosis that prevents the separation of the duplicated chromosomes (anaphase) until each chromosome is properly attached to the spindle. To achieve proper segregation, the two kinetochores on the sister chromatids must be attached to opposite spindle poles. Only this pattern of attachment will ensure that each daughter cell receives one copy of the chromosome. The defining biochemical feature of this checkpoint is the stimulation of the anaphase-promoting complex by M-phase cyclin-CDK complexes, which in turn causes the proteolytic destruction of cyclins and proteins that hold the sister chromatids together.

A kinetochore is a disc-shaped protein structure associated with duplicated chromatids in eukaryotic cells where the spindle fibers attach during cell division to pull sister chromatids apart. The kinetochore assembles on the centromere and links the chromosome to microtubule polymers from the mitotic spindle during mitosis and meiosis. Its proteins also help to hold the sister chromatids together and play a role in chromosome editing. Details of the specific areas of origin are unknown.
A spindle poison, also known as a spindle toxin, is a poison that disrupts cell division by affecting the protein threads that connect the centromere regions of chromosomes, known as spindles. Spindle poisons effectively cease the production of new cells by interrupting the mitosis phase of cell division at the spindle assembly checkpoint (SAC). However, as numerous and varied as they are, spindle poisons are not yet 100% effective at ending the formation of tumors (neoplasms). Although not 100% effective, substantive therapeutic efficacy has been found in these types of chemotherapeutic treatments. The mitotic spindle is composed of microtubules that aid, along with regulatory proteins; each other in the activity of appropriately segregating replicated chromosomes. Certain compounds affecting the mitotic spindle have proven highly effective against solid tumors and hematological malignancies.

Aurora B kinase is a protein that functions in the attachment of the mitotic spindle to the centromere.
Anaphase lag is a consequence of an event during cell division where sister chromatids do not properly separate from each other because of improper spindle formation. The chromosome or chromatid does not properly migrate during anaphase and the daughter cells will lose some genetic information. It is one of many causes of aneuploidy. This event can occur during both meiosis and mitosis with unique repercussions. In either case, anaphase lag will cause one daughter cell to receive a complete set of chromosomes while the other lacks one paired set of chromosomes, creating a form of monosomy. Whether the cell survives depends on which sister chromatid was lost and the background genomic state of the cell. The passage of abnormal numbers of chromosomes will have unique consequences with regards to mosaicism and development as well as the progression and heterogeneity of cancers.

Mitotic checkpoint serine/threonine-protein kinase BUB1 also known as BUB1 is an enzyme that in humans is encoded by the BUB1 gene.

Kinetochore protein NDC80 homolog is a protein that in humans is encoded by the NDC80 gene.

Centromere-associated protein E is a protein that in humans is encoded by the CENPE gene.

Mitotic checkpoint protein BUB3 is a protein that in humans is encoded by the BUB3 gene.

Kinetochore protein Nuf2 is a protein that in humans is encoded by the NUF2 gene.

An aster is a cellular structure shaped like a star, consisting of a centrosome and its associated microtubules during the early stages of mitosis in an animal cell. Asters do not form during mitosis in plants. Astral rays, composed of microtubules, radiate from the centrosphere and look like a cloud. Astral rays are one variant of microtubule which comes out of the centrosome; others include kinetochore microtubules and polar microtubules.

Mad1 is a non-essential protein which in yeast has a function in the spindle assembly checkpoint (SAC). This checkpoint monitors chromosome attachment to spindle microtubules and prevents cells from starting anaphase until the spindle is built up. The name Mad refers to the observation that mutant cells are mitotic arrest deficient (MAD) during microtubule depolymerization. Mad1 recruits the anaphase inhibitor Mad2 to unattached kinetochores and is essential for Mad2-Cdc20 complex formation in vivo but not in vitro. In vivo, Mad1 acts as a competitive inhibitor of the Mad2-Cdc20 complex. Mad1 is phosphorylated by Mps1 which then leads together with other activities to the formation of the mitotic checkpoint complex (MCC). Thereby it inhibits the activity of the anaphase-promoting complex/cyclosome (APC/C). Homologs of Mad1 are conserved in eukaryotes from yeast to mammals.
Biorientation is the phenomenon whereby microtubules emanating from different microtubule organizing centres (MTOCs) attach to kinetochores of sister chromatids. This results in the sister chromatids moving to opposite poles of the cell during cell division, and thus results in both daughter cells having the same genetic information.
In molecular biology, the protein domain named the Shugoshin N-terminal coiled-coil region is a domain found on the N-terminal region of the Shugoshin protein in eukaryotes. It has a role in attaching to the kinetochores, structures on the chromatids where microtubules attach. Shugoshin has a conserved coiled-coil N-terminal domain and a highly conserved C-terminal region. Shugoshin is a crucial target of Bub1 kinase that plays a central role in the cohesion of chromosomes during cell division.
Hesperadin is an aurora kinase inhibitor.
J. Richard McIntosh is a Distinguished Professor Emeritus in Molecular, Cellular, and Developmental Biology at the University of Colorado Boulder. McIntosh first graduated from Harvard with a BA in Physics in 1961, and again with a Ph.D. in Biophysics in 1968. He began his teaching career at Harvard but has spent most of his career at the University of Colorado Boulder. At the University of Colorado Boulder, McIntosh taught biology courses at both the undergraduate and graduate levels. Additionally, he created an undergraduate course in the biology of cancer towards the last several years of his teaching career. McIntosh's research career looks at a variety of things, including different parts of mitosis, microtubules, and motor proteins.
References
- ↑ "Examining chromosome-microtubule attachment". Archived from the original on 3 September 2010.
- ↑ London, Nitobe; Biggins, Sue (1 November 2014). "Signalling dynamics in the spindle checkpoint response". Nature Reviews Molecular Cell Biology. 15 (11): 736–748. doi:10.1038/nrm3888. PMC 4283840 . PMID 25303117.
- 1 2 "Molecular forces are key to proper cell division". www.sciencedaily.com. University of Massachusetts Amherst. January 21, 2013. Retrieved 3 February 2017.
- ↑ "CELLS Interactive Glossary: Syntelic attachment". bioscience.jbpub.com. Jones and Bartlett Publishers. Retrieved 3 February 2017.
- ↑ Walczak, Claire E.; Cai, Shang; Khodjakov, Alexey (20 January 2010). "Mechanisms of chromosome behaviour during mitosis". Nature Reviews Molecular Cell Biology. 11 (2): 91–102. doi:10.1038/nrm2832. PMC 2893392 . PMID 20068571.
- 1 2 Nicklas, R. Bruce (1997). "How Cells Get the Right Chromosomes". Science. 275 (5300): 632–637. doi:10.1126/science.275.5300.632. PMID 9005842.
- 1 2 Banerjee, Anand; Adames, Neil; Peccoud, Jean; Tyson, John J. (2020). "A stochastic model for error correction of kinetochore-microtubule attachments in budding yeast". PLOS One. 15 (8): e0236293. doi:10.1371/journal.pone.0236293. hdl: 10919/100303 . PMID 32760074.
- 1 2 3 4 Lončarek, Jadranka; Kisurina-Evgenieva, Olga; Vinogradova, Tatiana; Hergert, Polla; La Terra, Sabrina; Kapoor, Tarun M.; Khodjakov, Alexey (2007). "The centromere geometry essential for keeping mitosis error free is controlled by spindle forces". Nature. 450: 745–749. doi:10.1038/nature06344. PMC 2586812 . PMID 18046416.
- 1 2 Dewar, Hilary; Tanaka, Kozo; Nasmyth, Kim; Tanaka, Tomoyuki U. (2004). "Tension between two kinetochores suffices for their bi-orientation on the mitotic spindle". Nature. 428: 93–97. doi:10.1038/nature02328. PMID 14961024.
- 1 2 3 Storchová, Zuzana; Breneman, Amanda; Cande, Jessica; Dunn, Joshua; Burbank, Kendra; O'Toole, Eileen; Pellman, David (2006). "Genome-wide genetic analysis of polyploidy in yeast". Nature. 443: 541–547. doi:10.1038/nature05178. PMID 17024086.
- 1 2 3 4 5 6 Khodjakov, Alexey; Pines, Jonathon (2010). "Centromere tension: a divisive issue". Nature Cell Biology. 12 (10): 919–923. doi:10.1038/ncb1010-919. PMC 3052926 . PMID 20885417.
- 1 2 3 4 Lampson, Michael A.; Cheeseman, Iain M. (2011). "Sensing centromere tension: Aurora B and the regulation of kinetochore function". Trends in Cell Biology. 21 (3): 133–140. doi:10.1016/j.tcb.2010.10.007. hdl: 1721.1/98881 . PMID 21106376.
- 1 2 3 4 Tubman, Emily S.; Biggins, Sue; Odde, David J. (2017). "Stochastic Modeling Yields a Mechanistic Framework for Spindle Attachment Error Correction in Budding Yeast Mitosis". Cell Systems. 4 (6): 645-650.e5. doi:10.1016/j.cels.2017.05.003. PMC 5533192 . PMID 28601560.
- 1 2 Lampson, Michael A.; Grishchuk, Ekaterina L. (2017). "Mechanisms to Avoid and Correct Erroneous Kinetochore-Microtubule Attachments". Biology (Basel). 6 (1): 1. doi:10.3390/biology6010001. PMC 5371994 . PMID 28067761.
- 1 2 Thompson, Sarah L.; Compton, Duane A. (2011). "Chromosome missegregation in human cells arises through specific types of kinetochore–microtubule attachment errors". PNAS. 108 (44): 17974–17978. doi:10.1073/pnas.1109720108. PMC 3207692 . PMID 21997207.