Related Research Articles

Interferons are a group of signaling proteins made and released by host cells in response to the presence of several viruses. In a typical scenario, a virus-infected cell will release interferons causing nearby cells to heighten their anti-viral defenses.
Human embryonic kidney 293 cells, also often referred to as HEK 293, HEK-293, 293 cells, or less precisely as HEK cells, are a specific immortalised cell line originally derived from human embryonic kidney cells grown in tissue culture taken from a female fetus. HEK 293 cells have been widely used in cell biology research for many years, because of their reliable growth and propensity for transfection. They are also used by the biotechnology industry to produce therapeutic proteins and viruses for gene therapy.

Murine respirovirus, formerly Sendai virus (SeV) and previously also known as murine parainfluenza virus type 1 or hemagglutinating virus of Japan (HVJ), is an enveloped,150-200 nm in diameter, a negative sense, single-stranded RNA virus of the family Paramyxoviridae. It typically infects rodents and it is not pathogenic for humans or domestic animals. Sendai virus (SeV) is a member of genus Respirovirus. The virus was isolated in the city of Sendai in Japan in the early 1950s. Since then, it has been actively used in research as a model pathogen. The virus is infectious for many cancer cell lines, has oncolytic properties demonstrated in animal models and in naturally-occurring cancers in animals. SeV's ability to fuse eukaryotic cells and to form syncytium was used to produce hybridoma cells capable of manufacturing monoclonal antibodies in large quantities. Recent applications of SeV-based vectors include the reprogramming of somatic cells into induced pluripotent stem cells and vaccines creation. For vaccination purpose the Sendai virus-based constructs could be delivered in a form of nasal drops, which may be beneficial in inducing a mucosal immune response. SeV has several features that are important in a vector for a successful vaccine: the virus does not integrate into the host genome, it does not undergo genetic recombination, it replicates only in the cytoplasm without DNA intermediates or a nuclear phase and it is not causing any disease in humans or domestic animals. Sendai virus is used as a backbone for vaccine development against Mycobacterium tuberculosis that causes tuberculosis, against HIV-1 that causes AIDS and against respiratory syncytial virus (RSV) that causes respiratory infection in children. The vaccine development against Mycobacterium tuberculosis is in pre-clinical stage, against HIV-1 it reached phase II clinical trial and against RSV it is in phase I. Fudan University in collaboration with ID Pharma Co. Ltd. is engaged in development of the vaccine for COVID-19 prevention. SeV serves as a vaccine backbone vector in the project.

Interleukin-29 (IL-29) is a cytokine and it belongs to type III interferons group, also termed interferons λ (INF-λ). IL-29 plays an important role in the immune response against pathogenes and especially against viruses by mechanisms similar to type I interferons, but targeting primarily cells of epithelial origin and hepatocytes.

Interferon regulatory factor 3, also known as IRF3, is an interferon regulatory factor.

Signal transducer and activator of transcription 2 is a protein that in humans is encoded by the STAT2 gene. It is a member of the STAT protein family. This protein is critical to the biological response of type I interferons (IFNs). STAT2 sequence identity between mouse and human is only 68%.
NSP1, the product of rotavirus gene 5, is a nonstructural RNA-binding protein that contains a cysteine-rich region and is a component of early replication intermediates. RNA-folding predictions suggest that this region of the NSP1 mRNA can interact with itself, producing a stem-loop structure similar to that found near the 5'-terminus of the NSP1 mRNA.
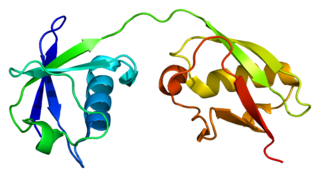
Interferon-stimulated gene 15 (ISG15) is a 17 kDA secreted protein that in humans is encoded by the ISG15 gene. ISG15 is induced by type I interferon (IFN) and serves many functions, acting both as an extracellular cytokine and an intracellular protein modifier. The precise functions are diverse and vary among species but include potentiation of Interferon gamma (IFN-II) production in lymphocytes, ubiquitin-like conjugation to newly-synthesized proteins and negative regulation of the IFN-I response.

RIG-I is a cytosolic pattern recognition receptor (PRR) responsible for the type-1 interferon (IFN1) response. RIG-I is an essential molecule in the innate immune system for recognizing cells that have been infected with a virus. These viruses can include West Nile virus, Japanese Encephalitis virus, influenza A, Sendai virus, flavivirus, and coronaviruses. RIG-I is structurally considered a helical ATP-dependent DExD/H box RNA helicase, that recognizes short viral double-stranded RNA (dsRNA) in the cytosol during a viral infection or other irregular RNAs. Once activated by the dsRNA, the N-terminus caspase activation and recruitment domains (CARDs) migrate and bind with CARDs attached to mitochondrial antiviral signaling protein (MAVS) to activate the signaling pathway for IFN1. IFN1s have three main functions: to limit the virus from spreading to nearby cells, promote an innate immune response, including inflammatory responses, and help activate the adaptive immune system. Other studies have shown that in different microenvironments, such as in cancerous cells, RIG-I has more functions other than viral recognition. RIG-I orthologs are found in mammals, geese, ducks, some fish, and some reptiles. RIG-I is in most cells, including various innate immune system cells, and is usually in an inactive state. Knockout mice that have been designed to have a deleted or non-functioning RIG-I gene are not healthy and typically die embryonically. If they survive, the mice have serious developmental dysfunction. The stimulator of interferon genes STING antagonizes RIG-1 by binding its N-terminus, probably as to avoid overactivation of RIG-1 signaling and the associated autoimmunity.

Interferon alpha-1 is a protein that in humans is encoded by the IFNA1 gene.

TBK1 is an enzyme with kinase activity. Specifically, it is a serine / threonine protein kinase. It is encoded by the TBK1 gene in humans. This kinase is mainly known for its role in innate immunity antiviral response. However, TBK1 also regulates cell proliferation, apoptosis, autophagy, and anti-tumor immunity. Insufficient regulation of TBK1 activity leads to autoimmune, neurodegenerative diseases or tumorogenesis.

Interferon alpha-2 is a protein that in humans is encoded by the IFNA2 gene.

Interferon regulatory factor 5 is a protein that in humans is encoded by the IRF5 gene. The IRF family is a group of transcription factors that are involved in signaling for virus responses in mammals along with regulation of certain cellular functions.

Mitochondrial antiviral-signaling protein (MAVS) is a protein that is essential for antiviral innate immunity. MAVS is located in the outer membrane of the mitochondria, peroxisomes, and endoplasmic reticulum (ER). Upon viral infection, a group of cytosolic proteins will detect the presence of the virus and bind to MAVS, thereby activating MAVS. The activation of MAVS leads the virally infected cell to secrete cytokines. This induces an immune response which kills the host's virally infected cells, resulting in clearance of the virus.
Baculovirus gene transfer into Mammalian cells, known from scientific research articles as BacMam, is the use of baculovirus to deliver genes to mammalian cells. Baculoviruses are insect cell viruses that can be modified to express proteins in mammalian cells. The unmodified baculovirus is able to enter mammalian cells, however its genes are not expressed unless a mammalian recognizable promoter is incorporated upstream of a gene of interest. Both unmodified baculovirus and baculovirus modified with a mammalian promoter (BacMam) are unable to replicate in humans and are thus non infectious.

MiR-155 is a microRNA that in humans is encoded by the MIR155 host gene or MIR155HG. MiR-155 plays a role in various physiological and pathological processes. Exogenous molecular control in vivo of miR-155 expression may inhibit malignant growth, viral infections, and enhance the progression of cardiovascular diseases.
RIG-like receptors are a type of intracellular pattern recognition receptor involved in the recognition of viruses by the innate immune system. RIG-I is the best characterized receptor within the RIG-I like receptor (RLR) family. Together with MDA5 and LGP2, this family of cytoplasmic pattern recognition receptors (PRRs) are sentinels for intracellular viral RNA that is a product of viral infection. The RLR receptors provide frontline defence against viral infections in most tissues.

Stimulator of interferon genes (STING), also known as transmembrane protein 173 (TMEM173) and MPYS/MITA/ERIS is a protein that in humans is encoded by the STING1 gene.
An interferon-stimulated gene (ISG) is a gene that can be expressed in response to stimulation by interferon. Interferons bind to receptors on the surface of a cell, initiating protein signaling pathways within the cell. This interaction leads to the expression of a subset of genes involved in the innate immune system response. ISGs are commonly expressed in response to viral infection, but also during bacterial infection and in the presence of parasites.
The cGAS–STING pathway is a component of the innate immune system that functions to detect the presence of cytosolic DNA and, in response, trigger expression of inflammatory genes that can lead to senescence or to the activation of defense mechanisms. DNA is normally found in the nucleus of the cell. Localization of DNA to the cytosol is associated with tumorigenesis, viral infection, and invasion by some intracellular bacteria. The cGAS – STING pathway acts to detect cytosolic DNA and induce an immune response.
References
- ↑ Sardiello, Marco; Cairo, Stefano; Fontanella, Bianca; Ballabio, Andrea; Meroni, Germana (2008-01-01). "Genomic analysis of the TRIM family reveals two groups of genes with distinct evolutionary properties". BMC Evolutionary Biology. 8: 225. doi:10.1186/1471-2148-8-225. ISSN 1471-2148. PMC 2533329 . PMID 18673550.
- ↑ Database, GeneCards Human Gene. "TRIM14 Gene - GeneCards | TRI14 Protein | TRI14 Antibody". www.genecards.org. Retrieved 2017-01-27.
- 1 2 Nenasheva, V. V.; Kovaleva, G. V.; Uryvaev, L. V.; Ionova, K. S.; Dedova, A. V.; Vorkunova, G. K.; Chernyshenko, S. V.; Khaidarova, N. V.; Tarantul, V. Z. (2015-05-07). "Enhanced expression of trim14 gene suppressed Sindbis virus reproduction and modulated the transcription of a large number of genes of innate immunity". Immunologic Research. 62 (3): 255–262. doi:10.1007/s12026-015-8653-1. ISSN 0257-277X. PMID 25948474. S2CID 27578364.
- 1 2 3 Chen, Meixin; Meng, Qingcai; Qin, Yunfei; Liang, Puping; Tan, Peng; He, Lian; Zhou, Yubin; Chen, Yongjun; Huang, Junjiu (2016). "TRIM14 Inhibits cGAS Degradation Mediated by Selective Autophagy Receptor p62 to Promote Innate Immune Responses". Molecular Cell. 64 (1): 105–119. doi: 10.1016/j.molcel.2016.08.025 . PMID 27666593.
- 1 2 Nenasheva, Valentina V.; Kovaleva, Galina V.; Khaidarova, Nella V.; Novosadova, Ekaterina V.; Manuilova, Ekaterina S.; Antonov, Stanislav A.; Tarantul, Vyacheslav Z. (2013-10-03). "Trim14 overexpression causes the same transcriptional changes in mouse embryonic stem cells and human HEK293 cells". In Vitro Cellular & Developmental Biology - Animal. 50 (2): 121–128. doi:10.1007/s11626-013-9683-4. ISSN 1071-2690. PMID 24092016. S2CID 9113530.
- ↑ Herquel, Benjamin; Ouararhni, Khalid; Khetchoumian, Konstantin; Ignat, Mihaela; Teletin, Marius; Mark, Manuel; Béchade, Guillaume; Dorsselaer, Alain Van; Sanglier-Cianférani, Sarah (2011-05-17). "Transcription cofactors TRIM24, TRIM28, and TRIM33 associate to form regulatory complexes that suppress murine hepatocellular carcinoma". Proceedings of the National Academy of Sciences. 108 (20): 8212–8217. Bibcode:2011PNAS..108.8212H. doi: 10.1073/pnas.1101544108 . ISSN 0027-8424. PMC 3100982 . PMID 21531907.
- ↑ Hirose, Satoshi; Nishizumi, Hirofumi; Sakano, Hitoshi (2003-11-14). "Pub, a novel PU.1 binding protein, regulates the transcriptional activity of PU.1". Biochemical and Biophysical Research Communications. 311 (2): 351–360. doi:10.1016/j.bbrc.2003.09.212. ISSN 0006-291X. PMID 14592421.
- ↑ Fisher, Robert C.; Scott, Edward W. (1998-01-01). "Role of PU.1 in Hematopoiesis". Stem Cells. 16 (1): 25–37. doi: 10.1002/stem.160025 . ISSN 1549-4918. PMID 9474745.
- ↑ Lloberas, J.; Soler, C.; Celada, A. (1999-04-01). "The key role of PU.1/SPI-1 in B cells, myeloid cells and macrophages". Immunology Today. 20 (4): 184–189. doi:10.1016/S0167-5699(99)01442-5. ISSN 0167-5699. PMID 10203717.
- ↑ Uchil, Pradeep D.; Quinlan, Brian D.; Chan, Wai-Tsing; Luna, Joseph M.; Mothes, Walther (2008-02-01). "TRIM E3 Ligases Interfere with Early and Late Stages of the Retroviral Life Cycle". PLOS Pathogens. 4 (2): e16. doi:10.1371/journal.ppat.0040016. ISSN 1553-7374. PMC 2222954 . PMID 18248090.
- ↑ Uchil, Pradeep D.; Hinz, Angelika; Siegel, Steven; Coenen-Stass, Anna; Pertel, Thomas; Luban, Jeremy; Mothes, Walther (2013-01-01). "TRIM Protein-Mediated Regulation of Inflammatory and Innate Immune Signaling and Its Association with Antiretroviral Activity". Journal of Virology. 87 (1): 257–272. doi:10.1128/JVI.01804-12. ISSN 0022-538X. PMC 3536418 . PMID 23077300.
- ↑ Zhou, Zhuo; Jia, Xue; Xue, Qinghua; Dou, Zhixun; Ma, Yijie; Zhao, Zhendong; Jiang, Zhengfan; He, Bin; Jin, Qi (2014-01-14). "TRIM14 is a mitochondrial adaptor that facilitates retinoic acid-inducible gene-I–like receptor-mediated innate immune response". Proceedings of the National Academy of Sciences. 111 (2): E245–E254. Bibcode:2014PNAS..111E.245Z. doi: 10.1073/pnas.1316941111 . ISSN 0027-8424. PMC 3896185 . PMID 24379373.
- 1 2 Wieland, Stefan; Thimme, Robert; Purcell, Robert H.; Chisari, Francis V. (2004-04-27). "Genomic analysis of the host response to hepatitis B virus infection". Proceedings of the National Academy of Sciences of the United States of America. 101 (17): 6669–6674. Bibcode:2004PNAS..101.6669W. doi: 10.1073/pnas.0401771101 . ISSN 0027-8424. PMC 404103 . PMID 15100412.
- ↑ Chen, Yin; Hamati, Edward; Lee, Pak-Kei; Lee, Wai-Ming; Wachi, Shinichiro; Schnurr, David; Yagi, Shigeo; Dolganov, Gregory; Boushey, Homer (2006-02-01). "Rhinovirus Induces Airway Epithelial Gene Expression through Double-Stranded RNA and IFN-Dependent Pathways". American Journal of Respiratory Cell and Molecular Biology. 34 (2): 192–203. doi:10.1165/rcmb.2004-0417OC. ISSN 1044-1549. PMC 2644182 . PMID 16210696.
- ↑ Ramilo, Octavio; Allman, Windy; Chung, Wendy; Mejias, Asuncion; Ardura, Monica; Glaser, Casey; Wittkowski, Knut M.; Piqueras, Bernard; Banchereau, Jacques (2007-03-01). "Gene expression patterns in blood leukocytes discriminate patients with acute infections". Blood. 109 (5): 2066–2077. doi:10.1182/blood-2006-02-002477. ISSN 0006-4971. PMC 1801073 . PMID 17105821.
- ↑ "Tissue expression of TRIM14 - Summary - The Human Protein Atlas". www.proteinatlas.org. Retrieved 2017-01-27.