
Proteasomes are protein complexes which degrade unneeded or damaged proteins by proteolysis, a chemical reaction that breaks peptide bonds. Enzymes that help such reactions are called proteases.

The thymus is a specialized primary lymphoid organ of the immune system. Within the thymus, thymus cell lymphocytes or T cells mature. T cells are critical to the adaptive immune system, where the body adapts to specific foreign invaders. The thymus is located in the upper front part of the chest, in the anterior superior mediastinum, behind the sternum, and in front of the heart. It is made up of two lobes, each consisting of a central medulla and an outer cortex, surrounded by a capsule.

T cells are one of the important types of white blood cells of the immune system and play a central role in the adaptive immune response. T cells can be distinguished from other lymphocytes by the presence of a T-cell receptor (TCR) on their cell surface.
Cross-presentation is the ability of certain professional antigen-presenting cells (mostly dendritic cells) to take up, process and present extracellular antigens with MHC class I molecules to CD8 T cells (cytotoxic T cells). Cross-priming, the result of this process, describes the stimulation of naive cytotoxic CD8+ T cells into activated cytotoxic CD8+ T cells. This process is necessary for immunity against most tumors and against viruses that infect dendritic cells and sabotage their presentation of virus antigens. Cross presentation is also required for the induction of cytotoxic immunity by vaccination with protein antigens, for example, tumour vaccination.
In immunology, central tolerance is the process of eliminating any developing T or B lymphocytes that are autoreactive, i.e. reactive to the body itself. Through elimination of autoreactive lymphocytes, tolerance ensures that the immune system does not attack self peptides. Lymphocyte maturation occurs in primary lymphoid organs such as the bone marrow and the thymus. In mammals, B cells mature in the bone marrow and T cells mature in the thymus.
A thymocyte is an immune cell present in the thymus, before it undergoes transformation into a T cell. Thymocytes are produced as stem cells in the bone marrow and reach the thymus via the blood.
MHC-restricted antigen recognition, or MHC restriction, refers to the fact that a T cell can interact with a self-major histocompatibility complex molecule and a foreign peptide bound to it, but will only respond to the antigen when it is bound to a particular MHC molecule.
An immunoproteasome is a type of proteasome that degrades ubiquitin-labeled proteins found in the cytoplasm in cells exposed to oxidative stress and proinflammatory stimuli. In general, proteasomes consist of a regulatory and a catalytic part. Immunoproteasomes are induced by interferon gamma and oxidative stress, which in the cell triggers the transcription of three catalytic subunits that do not occur in the classical proteasome. Another possible variation of proteasome is the thymoproteasome, which is located in the thymus and folds to present peptides to naive T cells.

The "C" sub-family of chemokine receptors contains only one member: XCR1, the receptor for XCL1 and XCL2.

Minor histocompatibility antigen are peptides presented on the cellular surface of donated organs that are known to give an immunological response in some organ transplants. They cause problems of rejection less frequently than those of the major histocompatibility complex (MHC). Minor histocompatibility antigens (MiHAs) are diverse, short segments of proteins and are referred to as peptides. These peptides are normally around 9-12 amino acids in length and are bound to both the major histocompatibility complex (MHC) class I and class II proteins. Peptide sequences can differ among individuals and these differences arise from SNPs in the coding region of genes, gene deletions, frameshift mutations, or insertions. About a third of the characterized MiHAs come from the Y chromosome. Prior to becoming a short peptide sequence, the proteins expressed by these polymorphic or diverse genes need to be digested in the proteasome into shorter peptides. These endogenous or self peptides are then transported into the endoplasmic reticulum with a peptide transporter pump called TAP where they encounter and bind to the MHC class I molecule. This contrasts with MHC class II molecules's antigens which are peptides derived from phagocytosis/endocytosis and molecular degradation of non-self entities' proteins, usually by antigen-presenting cells. MiHA antigens are either ubiquitously expressed in most tissue like skin and intestines or restrictively expressed in the immune cells.

Proteasome subunit beta type-10 as known as 20S proteasome subunit beta-2i is a protein that in humans is encoded by the PSMB10 gene.

Proteasome subunit beta type-8 as known as 20S proteasome subunit beta-5i is a protein that in humans is encoded by the PSMB8 gene. This protein is one of the 17 essential subunits that contributes to the complete assembly of 20S proteasome complex. In particular, proteasome subunit beta type-5, along with other beta subunits, assemble into two heptameric rings and subsequently a proteolytic chamber for substrate degradation. This protein contains "Chymotrypsin-like" activity and is capable of cleaving after large hydrophobic residues of peptide. The eukaryotic proteasome recognized degradable proteins, including damaged proteins for protein quality control purpose or key regulatory protein components for dynamic biological processes. The constitutive subunit beta1, beta2, and beta 5 can be replaced by their inducible counterparts beta1i, 2i, and 5i when cells are under the treatment of interferon-γ. The resulting proteasome complex becomes the so-called immunoproteasome. An essential function of the modified proteasome complex, the immunoproteasome, is the processing of numerous MHC class-I restricted T cell epitopes.

Proteasome subunit beta type-9 as known as 20S proteasome subunit beta-1i is a protein that in humans is encoded by the PSMB9 gene.
Proteasome subunit beta type-5 as known as 20S proteasome subunit beta-5 is a protein that in humans is encoded by the PSMB5 gene. This protein is one of the 17 essential subunits that contributes to the complete assembly of 20S proteasome complex. In particular, proteasome subunit beta type-5, along with other beta subunits, assemble into two heptameric rings and subsequently a proteolytic chamber for substrate degradation. This protein contains "chymotrypsin-like" activity and is capable of cleaving after large hydrophobic residues of peptide. The eukaryotic proteasome recognized degradable proteins, including damaged proteins for protein quality control purpose or key regulatory protein components for dynamic biological processes. An essential function of a modified proteasome, the immunoproteasome, is the processing of class I MHC peptides.

Forkhead box protein N1 is a protein that in humans is encoded by the FOXN1 gene.
Thymic nurse cells (TNCs) are large epithelial cells found in the cortex of the thymus and also in cortico-medullary junction. They have their own nucleus and are known to internalize thymocytes through extensions of plasma membrane. The cell surfaces of TNCs and their cytoplasmic vacuoles express MHC Class I and MHC Class II antigens. The interaction of these antigens with the developing thymocytes determines whether the thymocytes undergo positive or negative selection.
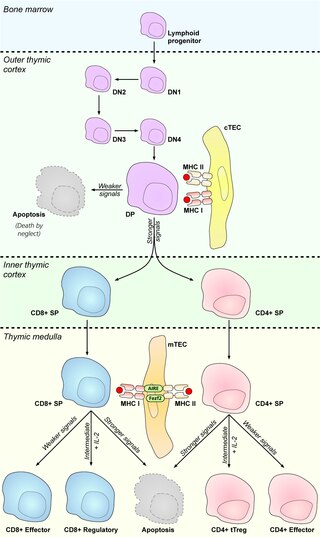
Medullary thymic epithelial cells (mTECs) represent a unique stromal cell population of the thymus which plays an essential role in the establishment of central tolerance. Therefore, mTECs rank among cells relevant for the development of functional mammal immune system.
Antigen transfer in the thymus is the transmission of self-antigens between thymic antigen-presenting cells which contributes to the establishment of T cell central tolerance.

Cortical thymic epithelial cells (cTECs) form unique parenchyma cell population of the thymus which critically contribute to the development of T cells.
Thymic epithelial cells (TECs) are specialized cells with high degree of anatomic, phenotypic and functional heterogeneity that are located in the outer layer (epithelium) of the thymic stroma. The thymus, as a primary lymphoid organ, mediates T cell development and maturation. The thymic microenvironment is established by TEC network filled with thymocytes in different developing stages. TECs and thymocytes are the most important components in the thymus, that are necessary for production of functionally competent T lymphocytes and self tolerance. Dysfunction of TECs causes several immunodeficiencies and autoimmune diseases.









