A phylogenetic tree, phylogeny or evolutionary tree is a graphical representation which shows the evolutionary history between a set of species or taxa during a specific time. In other words, it is a branching diagram or a tree showing the evolutionary relationships among various biological species or other entities based upon similarities and differences in their physical or genetic characteristics. In evolutionary biology, all life on Earth is theoretically part of a single phylogenetic tree, indicating common ancestry. Phylogenetics is the study of phylogenetic trees. The main challenge is to find a phylogenetic tree representing optimal evolutionary ancestry between a set of species or taxa. Computational phylogenetics focuses on the algorithms involved in finding optimal phylogenetic tree in the phylogenetic landscape.
The molecular clock is a figurative term for a technique that uses the mutation rate of biomolecules to deduce the time in prehistory when two or more life forms diverged. The biomolecular data used for such calculations are usually nucleotide sequences for DNA, RNA, or amino acid sequences for proteins.
The Archonta are a now-abandoned group of mammals, considered a superorder in some classifications, which consists of these orders:
A phylogenetic network is any graph used to visualize evolutionary relationships between nucleotide sequences, genes, chromosomes, genomes, or species. They are employed when reticulation events such as hybridization, horizontal gene transfer, recombination, or gene duplication and loss are believed to be involved. They differ from phylogenetic trees by the explicit modeling of richly linked networks, by means of the addition of hybrid nodes instead of only tree nodes. Phylogenetic trees are a subset of phylogenetic networks. Phylogenetic networks can be inferred and visualised with software such as SplitsTree, the R-package, phangorn, and, more recently, Dendroscope. A standard format for representing phylogenetic networks is a variant of Newick format which is extended to support networks as well as trees.
Computational phylogenetics, phylogeny inference, or phylogenetic inference focuses on computational and optimization algorithms, heuristics, and approaches involved in phylogenetic analyses. The goal is to find a phylogenetic tree representing optimal evolutionary ancestry between a set of genes, species, or taxa. Maximum likelihood, parsimony, Bayesian, and minimum evolution are typical optimality criteria used to assess how well a phylogenetic tree topology describes the sequence data. Nearest Neighbour Interchange (NNI), Subtree Prune and Regraft (SPR), and Tree Bisection and Reconnection (TBR), known as tree rearrangements, are deterministic algorithms to search for optimal or the best phylogenetic tree. The space and the landscape of searching for the optimal phylogenetic tree is known as phylogeny search space.
PHYLogeny Inference Package (PHYLIP) is a free computational phylogenetics package of programs for inferring evolutionary trees (phylogenies). It consists of 65 portable programs, i.e., the source code is written in the programming language C. As of version 3.696, it is licensed as open-source software; versions 3.695 and older were proprietary software freeware. Releases occur as source code, and as precompiled executables for many operating systems including Windows, Mac OS 8, Mac OS 9, OS X, Linux ; and FreeBSD from FreeBSD.org. Full documentation is written for all the programs in the package and is included therein. The programs in the phylip package were written by Professor Joseph Felsenstein, of the Department of Genome Sciences and the Department of Biology, University of Washington, Seattle.
In mathematics and phylogenetics, Newick tree format is a way of representing graph-theoretical trees with edge lengths using parentheses and commas. It was adopted by James Archie, William H. E. Day, Joseph Felsenstein, Wayne Maddison, Christopher Meacham, F. James Rohlf, and David Swofford, at two meetings in 1986, the second of which was at Newick's restaurant in Dover, New Hampshire, US. The adopted format is a generalization of the format developed by Meacham in 1984 for the first tree-drawing programs in Felsenstein's PHYLIP package.
The tree of life or universal tree of life is a metaphor, conceptual model, and research tool used to explore the evolution of life and describe the relationships between organisms, both living and extinct, as described in a famous passage in Charles Darwin's On the Origin of Species (1859).
The affinities of all the beings of the same class have sometimes been represented by a great tree. I believe this simile largely speaks the truth.

Eusporangiate ferns are vascular spore plants, whose sporangia arise from several epidermal cells and not from a single cell as in leptosporangiate ferns. Typically these ferns have reduced root systems and sporangia that produce large amounts of spores.

The Polypodiidae, commonly called leptosporangiate ferns, formerly Leptosporangiatae, are one of four subclasses of ferns, the largest of these being the largest group of living ferns, including some 11,000 species worldwide. The group has also been treated as the class Pteridopsida or Polypodiopsida, although other classifications assign them a different rank. Older names for the group include Filicidae and Filicales, although at least the "water ferns" were then treated separately.
The Pythonoidea, also known as pythonoid snakes, are a superfamily of snakes that contains pythons and other closely related python-like snakes. As of 2022, Pythonoidea contains 39 species, including the eponymous genus Python and 10 other genera of pythons, all in the family Pythonidae, as well as two lesser-known families, Loxocemidae and Xenopeltidae.

Terrabacteria is a taxon containing approximately two-thirds of prokaryote species, including those in the gram positive phyla as well as the phyla "Cyanobacteria", Chloroflexota, and Deinococcota.

Polypodiineae is a suborder of ferns in the order Polypodiales. It is equivalent to the clade eupolypods I in earlier systems, and to the very broadly defined family Polypodiaceae in the classification of Christenhusz & Chase (2014). It probably diverged from the suborder Aspleniineae during the mid-Cretaceous. The divergence is supported by both molecular data and an often overlooked morphological characteristic which lies in the vasculature of the petiole. Most species that make up the suborder have three vascular bundles. The only exceptions are the grammitid ferns which have one, and the genus Hypodematium which has two. This differs from eupolypods II which mostly have two vascular bundles.
A patrocladogram is a cladistic branching pattern that has been precisely modified by use of patristic distances ; a type of phylogram. The patristic distance is defined as, "the number of apomorphic step changes separating two taxa on a cladogram," and is used exclusively to determine the amount of divergence of a characteristic from a common ancestor. This means that cladistic and patristic distances are combined to construct a new tree using various phenetic algorithms. The purpose of the patrocladogram in biological classification is to form a hypothesis about which evolutionary processes are actually involved before making a taxonomic decision. Patrocladograms are based on biostatistics that include but are not limited to: parsimony, distance matrix, likelihood methods, and Bayesian probability. Some examples of genomically related data that can be used as inputs for these methods are: molecular sequences, whole genome sequences, gene frequencies, restriction sites, distance matrices, unique characters, mutations such as SNPs, and mitochondrial genome data.

Hydrobacteria is a taxon containing approximately one-third of prokaryote species, mostly gram-negative bacteria and their relatives. It was found to be the closest relative of an even larger group of Bacteria, Terrabacteria, which are mostly gram-positive bacteria. The name Hydrobacteria refers to the moist environment inferred for the common ancestor of those species. In contrast, species of Terrabacteria possess adaptations for life on land.

Stephen Blair Hedges is Laura H. Carnell Professor of Science and director of the Center for Biodiversity at Temple University where he researches the tree of life and leads conservation efforts in Haiti and elsewhere. He co-founded Haiti National Trust.
Triumph of the Vertebrates is a 2013 British documentary film by David Attenborough. It is about the evolution of vertebrates. The first part is From the Seas to the Skies, while the second is Dawn of the Mammals. The film uses a circular timetree of life generated by scientists S. Blair Hedges and Sudhir Kumar, from their TimeTree database, as a temporal framework for the production. The timetree was created using animated computer-generated imagery in scenes every 10 minutes during the 2-hour movie. The circular timetree was published by Hedges and Kumar in 2009 and Hedges was consulted during the production of the film.
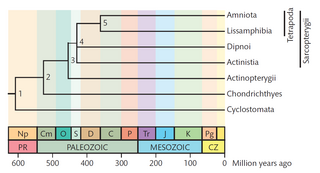
A timetree is a phylogenetic tree scaled to time. It shows the evolutionary relationships of a group of organisms in a temporal framework.

The Amerophidia, also known as amerophidian snakes, are a superfamily of snakes that contains two families: Aniliidae and the boa-like Tropidophiidae.

Afrophidia is a clade of alethinophidian snakes comprising the groups Henophidia and Caenophidia, essentially making up the snakes people commonly associate with. The name refers to the deep split between Afrophidia and their sister taxon, Amerophidia, which originated in South American origin, and the afrophidians was recently hypothesized to represent a vicariant event of the breakup of Gondwanan South America and Africa.












