Related Research Articles
Chromatin is a complex of DNA and protein found in eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important roles in reinforcing the DNA during cell division, preventing DNA damage, and regulating gene expression and DNA replication. During mitosis and meiosis, chromatin facilitates proper segregation of the chromosomes in anaphase; the characteristic shapes of chromosomes visible during this stage are the result of DNA being coiled into highly condensed chromatin.

In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei and in most Archaeal phyla. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn are wrapped into 30-nanometer fibers that form tightly packed chromatin. Histones prevent DNA from becoming tangled and protect it from DNA damage. In addition, histones play important roles in gene regulation and DNA replication. Without histones, unwound DNA in chromosomes would be very long. For example, each human cell has about 1.8 meters of DNA if completely stretched out; however, when wound about histones, this length is reduced to about 9 micrometers (0.09 mm) of 30 nm diameter chromatin fibers.

A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone proteins and resembles thread wrapped around a spool. The nucleosome is the fundamental subunit of chromatin. Each nucleosome is composed of a little less than two turns of DNA wrapped around a set of eight proteins called histones, which are known as a histone octamer. Each histone octamer is composed of two copies each of the histone proteins H2A, H2B, H3, and H4.
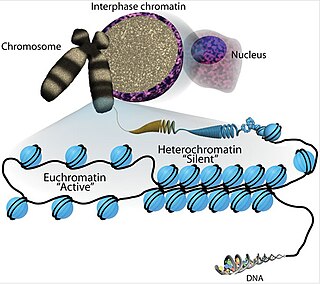
Euchromatin is a lightly packed form of chromatin that is enriched in genes, and is often under active transcription. Euchromatin stands in contrast to heterochromatin, which is tightly packed and less accessible for transcription. 92% of the human genome is euchromatic.

In molecular biology, a histone octamer is the eight-protein complex found at the center of a nucleosome core particle. It consists of two copies of each of the four core histone proteins. The octamer assembles when a tetramer, containing two copies of H3 and two of H4, complexes with two H2A/H2B dimers. Each histone has both an N-terminal tail and a C-terminal histone-fold. Each of these key components interacts with DNA in its own way through a series of weak interactions, including hydrogen bonds and salt bridges. These interactions keep the DNA and the histone octamer loosely associated, and ultimately allow the two to re-position or to separate entirely.
Histone H1 is one of the five main histone protein families which are components of chromatin in eukaryotic cells. Though highly conserved, it is nevertheless the most variable histone in sequence across species.

Histone H2A is one of the five main histone proteins involved in the structure of chromatin in eukaryotic cells.

The solenoid structure of chromatin is a model for the structure of the 30 nm fibre. It is a secondary chromatin structure which helps to package eukaryotic DNA into the nucleus.

Histone-modifying enzymes are enzymes involved in the modification of histone substrates after protein translation and affect cellular processes including gene expression. To safely store the eukaryotic genome, DNA is wrapped around four core histone proteins, which then join to form nucleosomes. These nucleosomes further fold together into highly condensed chromatin, which renders the organism's genetic material far less accessible to the factors required for gene transcription, DNA replication, recombination and repair. Subsequently, eukaryotic organisms have developed intricate mechanisms to overcome this repressive barrier imposed by the chromatin through histone modification, a type of post-translational modification which typically involves covalently attaching certain groups to histone residues. Once added to the histone, these groups elicit either a loose and open histone conformation, euchromatin, or a tight and closed histone conformation, heterochromatin. Euchromatin marks active transcription and gene expression, as the light packing of histones in this way allows entry for proteins involved in the transcription process. As such, the tightly packed heterochromatin marks the absence of current gene expression.
Chromatin remodeling is the dynamic modification of chromatin architecture to allow access of condensed genomic DNA to the regulatory transcription machinery proteins, and thereby control gene expression. Such remodeling is principally carried out by 1) covalent histone modifications by specific enzymes, e.g., histone acetyltransferases (HATs), deacetylases, methyltransferases, and kinases, and 2) ATP-dependent chromatin remodeling complexes which either move, eject or restructure nucleosomes. Besides actively regulating gene expression, dynamic remodeling of chromatin imparts an epigenetic regulatory role in several key biological processes, egg cells DNA replication and repair; apoptosis; chromosome segregation as well as development and pluripotency. Aberrations in chromatin remodeling proteins are found to be associated with human diseases, including cancer. Targeting chromatin remodeling pathways is currently evolving as a major therapeutic strategy in the treatment of several cancers.

Centromere protein A, also known as CENPA, is a protein which in humans is encoded by the CENPA gene. CENPA is a histone H3 variant which is the critical factor determining the kinetochore position(s) on each chromosome in most eukaryotes including humans.
FACT is a heterodimeric protein complex that affects eukaryotic RNA polymerase II transcription elongation both in vitro and in vivo. It was discovered in 1998 as a factor purified from human cells that was essential for productive, in vitro Pol II transcription on a chromatinized DNA template.

Daniela Bargellini Rhodes FRS is an Italian structural and molecular biologist. She was a senior scientist at the Laboratory of Molecular Biology in Cambridge, England, where she worked, and later studied for her PhD under the supervision of Nobel laureate Aaron Klug. Continuing her work under the tutelage of Aaron Klug at Cambridge, she was appointed group leader in 1983, obtained tenure in 1987 and was promoted to senior scientist in 1994. Subsequently, she served as director of studies between 2003 and 2006. She has also been visiting professor at both "La Sapienza" in Rome, Italy and the Rockefeller University in NY, USA.

H2A histone family, member B3 is a protein that in humans is encoded by the H2AFB3 gene.

Karolin Luger is an Austrian-American biochemist and biophysicist known for her work with nucleosomes and discovery of the three-dimensional structure of chromatin. She is a Distinguished Professor at the University of Colorado Boulder in the Biochemistry Department.
H3K4me3 is an epigenetic modification to the DNA packaging protein Histone H3 that indicates tri-methylation at the 4th lysine residue of the histone H3 protein and is often involved in the regulation of gene expression. The name denotes the addition of three methyl groups (trimethylation) to the lysine 4 on the histone H3 protein.
H3K56ac is an epigenetic modification to the DNA packaging protein Histone H3. It is a mark that indicates the acetylation at the 56th lysine residue of the histone H3 protein.

MNase-seq, short for micrococcal nuclease digestion with deep sequencing, is a molecular biological technique that was first pioneered in 2006 to measure nucleosome occupancy in the C. elegans genome, and was subsequently applied to the human genome in 2008. Though, the term ‘MNase-seq’ had not been coined until a year later, in 2009. Briefly, this technique relies on the use of the non-specific endo-exonuclease micrococcal nuclease, an enzyme derived from the bacteria Staphylococcus aureus, to bind and cleave protein-unbound regions of DNA on chromatin. DNA bound to histones or other chromatin-bound proteins may remain undigested. The uncut DNA is then purified from the proteins and sequenced through one or more of the various Next-Generation sequencing methods.
H3K36me is an epigenetic modification to the DNA packaging protein Histone H3, specifically, the mono-methylation at the 36th lysine residue of the histone H3 protein.
H3T45P is an epigenetic modification to the DNA packaging protein histone H3. It is a mark that indicates the phosphorylation the 45th threonine residue of the histone H3 protein.
References
- 1 2 3 4 "Timothy J. Richmond (membership number 2024)". Academia Europaea.
- 1 2 3 4 "Neue Mitglieder der Leopoldina 2004" (PDF). Deutsche Akademie der Naturforscher Leopoldina. p. 70.
- ↑ "2010 Bios". Yale Graduate School of Arts and Sciences.
- ↑ "Richmond, Timothy J., Prof. Dr". ETH Zürich.
- ↑ "Timothy J. Richmond, Ph.D." Chemistry Tree (academictree.org).
- ↑ "Historic Fellows". American Association for the Advancement of Science (aaas.org).
- ↑ "EMBO Member Timothy J. Richmond". EMBO.
- ↑ "Member Directory: Timothy J. Richmond". National Academy of Sciences.
- ↑ "Timothy Richmond". Purdue to award 15 honorary doctorates, Purdue News. April 2001.
- ↑ "Professor Timothy J. Richmond, Winner of the 2002 Louis-Jeantet Prize for medicine". Fondation Louis-Jeantet. October 2017.
- ↑ "Laureates". Marcel Benoist Foundation.
- ↑ Laureates of the 2023 WLA Prize Announced, The WLA Prize