Related Research Articles
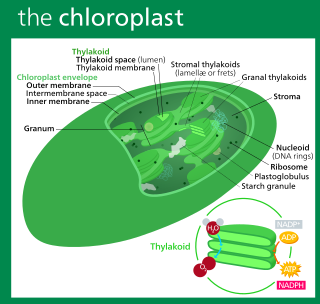
A chloroplast is a type of membrane-bound organelle known as a plastid that conducts photosynthesis mostly in plant and algal cells. The photosynthetic pigment chlorophyll captures the energy from sunlight, converts it, and stores it in the energy-storage molecules ATP and NADPH while freeing oxygen from water in the cells. The ATP and NADPH is then used to make organic molecules from carbon dioxide in a process known as the Calvin cycle. Chloroplasts carry out a number of other functions, including fatty acid synthesis, amino acid synthesis, and the immune response in plants. The number of chloroplasts per cell varies from one, in unicellular algae, up to 100 in plants like Arabidopsis and wheat.

The endoplasmic reticulum (ER) is, in essence, the transportation system of the eukaryotic cell, and has many other important functions such as protein folding. It is a type of organelle made up of two subunits – rough endoplasmic reticulum (RER), and smooth endoplasmic reticulum (SER). The endoplasmic reticulum is found in most eukaryotic cells and forms an interconnected network of flattened, membrane-enclosed sacs known as cisternae, and tubular structures in the SER. The membranes of the ER are continuous with the outer nuclear membrane. The endoplasmic reticulum is not found in red blood cells, or spermatozoa.
Protein targeting or protein sorting is the biological mechanism by which proteins are transported to their appropriate destinations within or outside the cell. Proteins can be targeted to the inner space of an organelle, different intracellular membranes, the plasma membrane, or to the exterior of the cell via secretion. Information contained in the protein itself directs this delivery process. Correct sorting is crucial for the cell; errors or dysfunction in sorting have been linked to multiple diseases.
A transmembrane domain (TMD) is a membrane-spanning protein domain. TMDs generally adopt an alpha helix topological conformation, although some TMDs such as those in porins can adopt a different conformation. Because the interior of the lipid bilayer is hydrophobic, the amino acid residues in TMDs are often hydrophobic, although proteins such as membrane pumps and ion channels can contain polar residues. TMDs vary greatly in length, sequence, and hydrophobicity, adopting organelle-specific properties.

A transmembrane protein (TP) is a type of integral membrane protein that spans the entirety of the cell membrane. Many transmembrane proteins function as gateways to permit the transport of specific substances across the membrane. They frequently undergo significant conformational changes to move a substance through the membrane. They are usually highly hydrophobic and aggregate and precipitate in water. They require detergents or nonpolar solvents for extraction, although some of them (beta-barrels) can be also extracted using denaturing agents.

Peripheral membrane proteins, or extrinsic membrane proteins, are membrane proteins that adhere only temporarily to the biological membrane with which they are associated. These proteins attach to integral membrane proteins, or penetrate the peripheral regions of the lipid bilayer. The regulatory protein subunits of many ion channels and transmembrane receptors, for example, may be defined as peripheral membrane proteins. In contrast to integral membrane proteins, peripheral membrane proteins tend to collect in the water-soluble component, or fraction, of all the proteins extracted during a protein purification procedure. Proteins with GPI anchors are an exception to this rule and can have purification properties similar to those of integral membrane proteins.
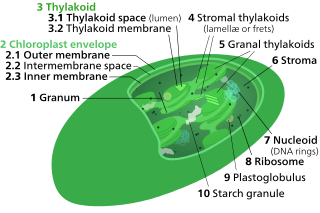
Thylakoids are membrane-bound compartments inside chloroplasts and cyanobacteria. They are the site of the light-dependent reactions of photosynthesis. Thylakoids consist of a thylakoid membrane surrounding a thylakoid lumen. Chloroplast thylakoids frequently form stacks of disks referred to as grana. Grana are connected by intergranal/stromal thylakoids, which join granum stacks together as a single functional compartment.
The signal recognition particle (SRP) is an abundant, cytosolic, universally conserved ribonucleoprotein that recognizes and targets specific proteins to the endoplasmic reticulum in eukaryotes and the plasma membrane in prokaryotes.
A signal peptide is a short peptide present at the N-terminus of most newly synthesized proteins that are destined toward the secretory pathway. These proteins include those that reside either inside certain organelles, secreted from the cell, or inserted into most cellular membranes. Although most type I membrane-bound proteins have signal peptides, the majority of type II and multi-spanning membrane-bound proteins are targeted to the secretory pathway by their first transmembrane domain, which biochemically resembles a signal sequence except that it is not cleaved. They are a kind of target peptide.
The translocon is a complex of proteins associated with the translocation of polypeptides across membranes. In eukaryotes the term translocon most commonly refers to the complex that transports nascent polypeptides with a targeting signal sequence into the interior space of the endoplasmic reticulum (ER) from the cytosol. This translocation process requires the protein to cross a hydrophobic lipid bilayer. The same complex is also used to integrate nascent proteins into the membrane itself. In prokaryotes, a similar protein complex transports polypeptides across the (inner) plasma membrane or integrates membrane proteins. In either case, the protein complex are formed from Sec proteins, with the heterotrimeric Sec61 being the channel. In prokaryotes, the homologous channel complex is known as SecYEG.
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the amine group is bonded to the carboxylic group of another amino acid, making it a chain. That leaves a free carboxylic group at one end of the peptide, called the C-terminus, and a free amine group on the other end called the N-terminus. By convention, peptide sequences are written N-terminus to C-terminus, left to right (in LTR writing systems). This correlates the translation direction to the text direction, because when a protein is translated from messenger RNA, it is created from the N-terminus to the C-terminus, as amino acids are added to the carboxyl end of the protein.
In cell biology, microsomes are heterogeneous vesicle-like artifacts re-formed from pieces of the endoplasmic reticulum (ER) when eukaryotic cells are broken-up in the laboratory; microsomes are not present in healthy, living cells.
Sec61, termed SecYEG in prokaryotes, is a membrane protein complex found in all domains of life. As the core component of the translocon, it transports proteins to the endoplasmic reticulum in eukaryotes and out of the cell in prokaryotes. It is a doughnut-shaped pore through the membrane with 3 different subunits (heterotrimeric), SecY (α), SecE (γ), and SecG (β). It has a region called the plug that blocks transport into or out of the ER. This plug is displaced when the hydrophobic region of a nascent polypeptide interacts with another region of Sec61 called the seam, allowing translocation of the polypeptide into the ER lumen.

Signal recognition particle (SRP) receptor, also called the docking protein, is a dimer composed of 2 different subunits that are associated exclusively with the rough ER in mammalian cells. Its main function is to identify the SRP units. SRP is a molecule that helps the ribosome-mRNA-polypeptide complexes to settle down on the membrane of the endoplasmic reticulum.
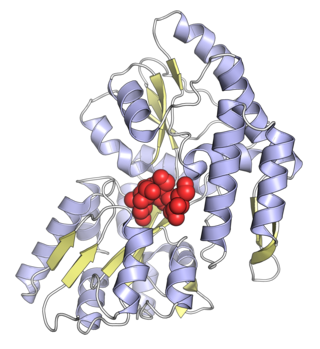
Maltose-binding protein (MBP) is a part of the maltose/maltodextrin system of Escherichia coli, which is responsible for the uptake and efficient catabolism of maltodextrins. It is a complex regulatory and transport system involving many proteins and protein complexes. MBP has an approximate molecular mass of 42.5 kilodaltons.

The signal recognition particle RNA, is part of the signal recognition particle (SRP) ribonucleoprotein complex. SRP recognizes the signal peptide and binds to the ribosome, halting protein synthesis. SRP-receptor is a protein that is embedded in a membrane, and which contains a transmembrane pore. When the SRP-ribosome complex binds to SRP-receptor, SRP releases the ribosome and drifts away. The ribosome resumes protein synthesis, but now the protein is moving through the SRP-receptor transmembrane pore.
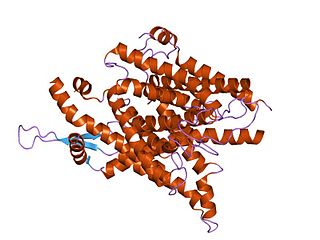
The SecY protein is the main transmembrane subunit of the bacterial Sec export pathway and of a protein-secreting ATPase complex, also known as a SecYEG translocon. Homologs of the SecYEG complex are found in eukaryotes and in archaea, where the subunit is known as Sec61α.
Chloroplast DNA (cpDNA) is the DNA located in chloroplasts, which are photosynthetic organelles located within the cells of some eukaryotic organisms. Chloroplasts, like other types of plastid, contain a genome separate from that in the cell nucleus. The existence of chloroplast DNA was identified biochemically in 1959, and confirmed by electron microscopy in 1962. The discoveries that the chloroplast contains ribosomes and performs protein synthesis revealed that the chloroplast is genetically semi-autonomous. The first complete chloroplast genome sequences were published in 1986, Nicotiana tabacum (tobacco) by Sugiura and colleagues and Marchantia polymorpha (liverwort) by Ozeki et al. Since then, a great number of chloroplast DNAs from various species have been sequenced.
A target peptide is a short peptide chain that directs the transport of a protein to a specific region in the cell, including the nucleus, mitochondria, endoplasmic reticulum (ER), chloroplast, apoplast, peroxisome and plasma membrane. Some target peptides are cleaved from the protein by signal peptidases after the proteins are transported.
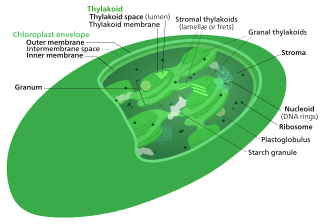
The TIC and TOC complexes are translocons located in the chloroplast of a eukaryotic cell, that is, protein complexes that facilitate the transfer of proteins in and out through the chloroplast's membrane. It mainly transports proteins made in the cytoplasm into the chloroplast. The TIC complex(translocon on the inner chloroplast membrane) is located in the inner envelope of the chloroplast. The TOC complex(translocon on the outer chloroplast membrane) is located in the outer envelope of the chloroplast.
References
- ↑ Robinson, A; Austen, B (1 September 1987). "The role of topogenic sequences in the movement of proteins through membranes". Biochem. J. 246 (2): 249–261. doi:10.1042/bj2460249. PMC 1148271 . PMID 3318806.
- 1 2 Gould, Sven; Waller, R; McFadden, G (June 2008). "Plastid Evolution". Annual Review of Plant Biology. 59: 491–517. doi:10.1146/annurev.arplant.59.032607.092915. PMID 18315522.
- ↑ Lodish, Harvey (2013). Molecular Cell Biology. Macmillan Higher Education. pp. 587–588. ISBN 978-1-4292-3413-9.