
Transcription is the process of copying a segment of DNA into RNA. The segments of DNA transcribed into RNA molecules that can encode proteins are said to produce messenger RNA (mRNA). Other segments of DNA are copied into RNA molecules called non-coding RNAs (ncRNAs). Averaged over multiple cell types in a given tissue, the quantity of mRNA is more than 10 times the quantity of ncRNA. The general preponderance of mRNA in cells is valid even though less than 2% of the human genome can be transcribed into mRNA, while at least 80% of mammalian genomic DNA can be actively transcribed, with the majority of this 80% considered to be ncRNA.
In genetics, an enhancer is a short region of DNA that can be bound by proteins (activators) to increase the likelihood that transcription of a particular gene will occur. These proteins are usually referred to as transcription factors. Enhancers are cis-acting. They can be located up to 1 Mbp away from the gene, upstream or downstream from the start site. There are hundreds of thousands of enhancers in the human genome. They are found in both prokaryotes and eukaryotes.

Oct-4, also known as POU5F1, is a protein that in humans is encoded by the POU5F1 gene. Oct-4 is a homeodomain transcription factor of the POU family. It is critically involved in the self-renewal of undifferentiated embryonic stem cells. As such, it is frequently used as a marker for undifferentiated cells. Oct-4 expression must be closely regulated; too much or too little will cause differentiation of the cells.
Myc is a family of regulator genes and proto-oncogenes that code for transcription factors. The Myc family consists of three related human genes: c-myc (MYC), l-myc (MYCL), and n-myc (MYCN). c-myc was the first gene to be discovered in this family, due to homology with the viral gene v-myc.

N-myc proto-oncogene protein also known as N-Myc or basic helix-loop-helix protein 37 (bHLHe37), is a protein that in humans is encoded by the MYCN gene.

Induced pluripotent stem cells are a type of pluripotent stem cell that can be generated directly from a somatic cell. The iPSC technology was pioneered by Shinya Yamanaka’s lab in Kyoto, Japan, who showed in 2006 that the introduction of four specific genes, collectively known as Yamanaka factors, encoding transcription factors could convert somatic cells into pluripotent stem cells. He was awarded the 2012 Nobel Prize along with Sir John Gurdon "for the discovery that mature cells can be reprogrammed to become pluripotent."

Cell division protein kinase 8 is an enzyme that in humans is encoded by the CDK8 gene.

Nuclear transcription factor Y subunit gamma is a protein that in humans is encoded by the NFYC gene.

T-box transcription factor TBX3 is a protein that in humans is encoded by the TBX3 gene.

L-myc-1 proto-oncogene protein is a protein that in humans is encoded by the MYCL1 gene.

Sal-like protein 4(SALL4) is a transcription factor encoded by a member of the Spalt-like (SALL) gene family, SALL4. The SALL genes were identified based on their sequence homology to Spalt, which is a homeotic gene originally cloned in Drosophila melanogaster that is important for terminal trunk structure formation in embryogenesis and imaginal disc development in the larval stages. There are four human SALL proteins with structural homology and playing diverse roles in embryonic development, kidney function, and cancer. The SALL4 gene encodes at least three isoforms, termed A, B, and C, through alternative splicing, with the A and B forms being the most studied. SALL4 can alter gene expression changes through its interaction with many co-factors and epigenetic complexes. It is also known as a key embryonic stem cell (ESC) factor.

Bromodomain-containing protein 4 is a protein that in humans is encoded by the BRD4 gene.

Bromodomain-containing protein 3 (BRD3) also known as RING3-like protein (RING3L) is a protein that in humans is encoded by the BRD3 gene. This gene was identified based on its homology to the gene encoding the RING3 (BRD2) protein, a serine/threonine kinase. The gene maps to 9q34, a region which contains several major histocompatibility complex (MHC) genes.

General transcription factor IIF subunit 1 is a protein that in humans is encoded by the GTF2F1 gene.

Alvocidib is a flavonoid alkaloid CDK9 kinase inhibitor under clinical development by Tolero Pharmaceuticals for the treatment of acute myeloid leukemia. It has been studied also for the treatment of arthritis and atherosclerotic plaque formation. The target of alvocidib is the positive transcription elongation factor P-TEFb. Treatment of cells with alvocidib leads to inhibition of P-TEFb and the loss of mRNA production.

JQ1 is a thienotriazolodiazepine and a potent inhibitor of the BET family of bromodomain proteins which include BRD2, BRD3, BRD4, and the testis-specific protein BRDT in mammals. BET inhibitors structurally similar to JQ1 are being tested in clinical trials for a variety of cancers including NUT midline carcinoma. It was developed by the James Bradner laboratory at Brigham and Women's Hospital and named after chemist Jun Qi. The chemical structure was inspired by patent of similar BET inhibitors by Mitsubishi Tanabe Pharma [WO/2009/084693]. Structurally it is related to benzodiazepines. While widely used in laboratory applications, JQ1 is not itself being used in human clinical trials because it has a short half life.

Glis1 is gene encoding a Krüppel-like protein of the same name whose locus is found on Chromosome 1p32.3. The gene is enriched in unfertilised eggs and embryos at the one cell stage and it can be used to promote direct reprogramming of somatic cells to induced pluripotent stem cells, also known as iPS cells. Glis1 is a highly promiscuous transcription factor, regulating the expression of numerous genes, either positively or negatively. In organisms, Glis1 does not appear to have any directly important functions. Mice whose Glis1 gene has been removed have no noticeable change to their phenotype.
BET inhibitors are a class of drugs that reversibly bind the bromodomains of Bromodomain and Extra-Terminal motif (BET) proteins BRD2, BRD3, BRD4, and BRDT, and prevent protein-protein interaction between BET proteins and acetylated histones and transcription factors.
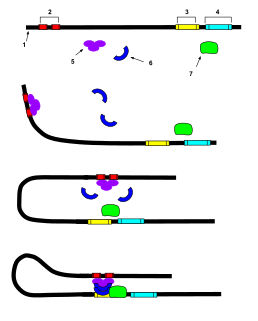
In genetics, a super-enhancer is a region of the mammalian genome comprising multiple enhancers that is collectively bound by an array of transcription factor proteins to drive transcription of genes involved in cell identity. Because super-enhancers are frequently identified near genes important for controlling and defining cell identity, they may thus be used to quickly identify key nodes regulating cell identity.

Richard Allen Young is an American geneticist, a Member of Whitehead Institute, and a professor of biology at the Massachusetts Institute of Technology. He is a pioneer in the systems biology of gene control who has developed genomics technologies and concepts key to understanding gene control in human health and disease. He has served as an advisor to the World Health Organization and the National Institutes of Health. He is a member of the National Academy of Sciences and the National Academy of Medicine. Scientific American has recognized him as one of the top 50 leaders in science, technology and business. Young is among the most Highly Cited Researchers in his field.
















