
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term exon refers to both the DNA sequence within a gene and to the corresponding sequence in RNA transcripts. In RNA splicing, introns are removed and exons are covalently joined to one another as part of generating the mature RNA. Just as the entire set of genes for a species constitutes the genome, the entire set of exons constitutes the exome.

In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.

In molecular genetics, the three prime untranslated region (3′-UTR) is the section of messenger RNA (mRNA) that immediately follows the translation termination codon. The 3′-UTR often contains regulatory regions that post-transcriptionally influence gene expression.
The 5′ untranslated region is the region of a messenger RNA (mRNA) that is directly upstream from the initiation codon. This region is important for the regulation of translation of a transcript by differing mechanisms in viruses, prokaryotes and eukaryotes. While called untranslated, the 5′ UTR or a portion of it is sometimes translated into a protein product. This product can then regulate the translation of the main coding sequence of the mRNA. In many organisms, however, the 5′ UTR is completely untranslated, instead forming a complex secondary structure to regulate translation.

Directionality, in molecular biology and biochemistry, is the end-to-end chemical orientation of a single strand of nucleic acid. In a single strand of DNA or RNA, the chemical convention of naming carbon atoms in the nucleotide pentose-sugar-ring means that there will be a 5′ end, which frequently contains a phosphate group attached to the 5′ carbon of the ribose ring, and a 3′ end, which typically is unmodified from the ribose -OH substituent. In a DNA double helix, the strands run in opposite directions to permit base pairing between them, which is essential for replication or transcription of the encoded information.
The Let-7 microRNA precursor was identified from a study of developmental timing in C. elegans, and was later shown to be part of a much larger class of non-coding RNAs termed microRNAs. miR-98 microRNA precursor from human is a let-7 family member. Let-7 miRNAs have now been predicted or experimentally confirmed in a wide range of species (MIPF0000002). miRNAs are initially transcribed in long transcripts called primary miRNAs (pri-miRNAs), which are processed in the nucleus by Drosha and Pasha to hairpin structures of about 70 nucleotide. These precursors (pre-miRNAs) are exported to the cytoplasm by exportin5, where they are subsequently processed by the enzyme Dicer to a ~22 nucleotide mature miRNA. The involvement of Dicer in miRNA processing demonstrates a relationship with the phenomenon of RNA interference.

In molecular biology lin-4 is a microRNA (miRNA) that was identified from a study of developmental timing in the nematode Caenorhabditis elegans. It was the first to be discovered of the miRNAs, a class of non-coding RNAs involved in gene regulation. miRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a 21 nucleotide product. The extents of the hairpin precursors are not generally known and are estimated based on hairpin prediction. The products are thought to have regulatory roles through complete or partial complementarity to mRNA. The lin-4 gene has been found to lie within a 4.11kb intron of a separate host gene.
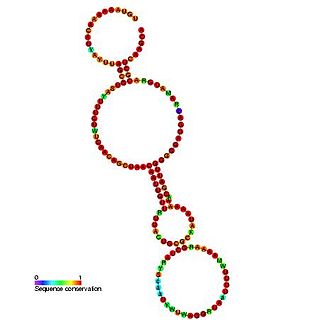
The PrfA thermoregulator UTR is an RNA thermometer found in the 5' UTR of the prfA gene. In Listeria monocytogenes, virulence genes are maximally expressed at 37 °C but are almost silent at 30 °C. The genes are controlled by PrfA, a transcriptional activator whose expression is thermoregulated. It has been shown that the untranslated mRNA (UTR) preceding prfA, forms a secondary structure, which masks the ribosome binding region. It is thought that at 37 °C, the hairpin structure 'melts' and the SD sequence is unmasked.

In molecular genetics, an untranslated region refers to either of two sections, one on each side of a coding sequence on a strand of mRNA. If it is found on the 5' side, it is called the 5' UTR, or if it is found on the 3' side, it is called the 3' UTR. mRNA is RNA that carries information from DNA to the ribosome, the site of protein synthesis (translation) within a cell. The mRNA is initially transcribed from the corresponding DNA sequence and then translated into protein. However, several regions of the mRNA are usually not translated into protein, including the 5' and 3' UTRs.
Gary Bruce Ruvkun is an American molecular biologist at Massachusetts General Hospital and professor of genetics at Harvard Medical School in Boston. Ruvkun discovered the mechanism by which lin-4, the first microRNA (miRNA) discovered by Victor Ambros, regulates the translation of target messenger RNAs via imperfect base-pairing to those targets, and discovered the second miRNA, let-7, and that it is conserved across animal phylogeny, including in humans. These miRNA discoveries revealed a new world of RNA regulation at an unprecedented small size scale, and the mechanism of that regulation. Ruvkun also discovered many features of insulin-like signaling in the regulation of aging and metabolism. He was elected a Member of the American Philosophical Society in 2019.
An upstream open reading frame (uORF) is an open reading frame (ORF) within the 5' untranslated region (5'UTR) of an mRNA. uORFs can regulate eukaryotic gene expression. Translation of the uORF typically inhibits downstream expression of the primary ORF. However, in some genes such as yeast GCN4, translation of specific uORFs may increase translation of the main ORF. In bacteria, uORFs are called leader peptides and were originally discovered on the basis of their impact on the regulation of genes involved in the synthesis or transport of amino acids.

Red clover necrotic mosaic virus (RCNMV) contains several structural elements present within the 3′ and 5′ untranslated regions (UTR) of the genome that enhance translation. In eukaryotes transcription is a prerequisite for translation. During transcription the pre-mRNA transcript is processes where a 5′ cap is attached onto mRNA and this 5′ cap allows for ribosome assembly onto the mRNA as it acts as a binding site for the eukaryotic initiation factor eIF4F. Once eIF4F is bound to the mRNA this protein complex interacts with the poly(A) binding protein which is present within the 3′ UTR and results in mRNA circularization. This multiprotein-mRNA complex then recruits the ribosome subunits and scans the mRNA until it reaches the start codon. Transcription of viral genomes differs from eukaryotes as viral genomes produce mRNA transcripts that lack a 5’ cap site. Despite lacking a cap site viral genes contain a structural element within the 5’ UTR known as an internal ribosome entry site (IRES). IRES is a structural element that recruits the 40s ribosome subunit to the mRNA within close proximity of the start codon.

Barley yellow dwarf virus 5' UTR is a non-coding RNA element containing structural elements required for translation of the genome of the plant disease pathogen Barley yellow dwarf virus.
The Atlas of UTR Regulatory Activity (AURA), a biological database, now at its second version, is a manually curated and comprehensive catalog of human 5' and 3' untranslated sequences (UTR) and UTR regulatory annotations. It includes basic annotation, phylogenetic conservation, binding sites for RNA-binding proteins and miRNA, cis-elements, RNA methylation and editing data, and more, for human and mouse. Through its intuitive web interface, it furthermore provides full access to a wealth of information that integrates RNA sequence and structure data, variation sites, gene synteny, gene and protein expression and gene functional descriptions from scientific literature and specialized databases. Eventually, it provides several tool for batch analysis of gene lists, allowing the tracing of post-transcriptional regulatory networks.
lsy-6 microRNA belongs to the class of miRNAs; these function to regulate the expression levels of other genes by several mechanisms. lsy-6 is a short non-coding RNA molecule and the first miRNA identified as having a role in nervous system development. It regulates left-right neuronal asymmetry in the nematode worm Caenorhabditis elegans.
In bioinformatics, TargetScan is a web server that predicts biological targets of microRNAs (miRNAs) by searching for the presence of sites that match the seed region of each miRNA. For many species, other types of sites, known as 3'-compensatory sites are also identified. These miRNA target predictions are regularly updated and improved by the laboratory of David Bartel in conjunction with the Whitehead Institute Bioinformatics and Research Computing Group.
WormBase is an online biological database about the biology and genome of the nematode model organism Caenorhabditis elegans and contains information about other related nematodes. WormBase is used by the C. elegans research community both as an information resource and as a place to publish and distribute their results. The database is regularly updated with new versions being released every two months. WormBase is one of the organizations participating in the Generic Model Organism Database (GMOD) project.

Flavivirus 5' UTR are untranslated regions in the genome of viruses in the genus Flavivirus.
Flavivirus 3' UTR are untranslated regions in the genome of viruses in the genus Flavivirus.








