Related Research Articles

In genetics, a promoter is a sequence of DNA to which proteins bind to initiate transcription of a single RNA transcript from the DNA downstream of the promoter. The RNA transcript may encode a protein (mRNA), or can have a function in and of itself, such as tRNA or rRNA. Promoters are located near the transcription start sites of genes, upstream on the DNA . Promoters can be about 100–1000 base pairs long, the sequence of which is highly dependent on the gene and product of transcription, type or class of RNA polymerase recruited to the site, and species of organism.

A non-coding RNA (ncRNA) is a functional RNA molecule that is not translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally important types of non-coding RNAs include transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), as well as small RNAs such as microRNAs, siRNAs, piRNAs, snoRNAs, snRNAs, exRNAs, scaRNAs and the long ncRNAs such as Xist and HOTAIR.

The CpG sites or CG sites are regions of DNA where a cytosine nucleotide is followed by a guanine nucleotide in the linear sequence of bases along its 5' → 3' direction. CpG sites occur with high frequency in genomic regions called CpG islands.

Pseudogenes are nonfunctional segments of DNA that resemble functional genes. Most arise as superfluous copies of functional genes, either directly by gene duplication or indirectly by reverse transcription of an mRNA transcript. Pseudogenes are usually identified when genome sequence analysis finds gene-like sequences that lack regulatory sequences needed for transcription or translation, or whose coding sequences are obviously defective due to frameshifts or premature stop codons. Pseudogenes are a type of junk DNA.
An Alu element is a short stretch of DNA originally characterized by the action of the Arthrobacter luteus (Alu) restriction endonuclease. Alu elements are the most abundant transposable elements in the human genome, present in excess of one million copies. Alu elements were thought to be selfish or parasitic DNA, because their sole known function is self reproduction. However, they are likely to play a role in evolution and have been used as genetic markers. They are derived from the small cytoplasmic 7SL RNA, a component of the signal recognition particle. Alu elements are highly conserved within primate genomes and originated in the genome of an ancestor of Supraprimates.

In evolutionary biology, conserved sequences are identical or similar sequences in nucleic acids or proteins across species, or within a genome, or between donor and receptor taxa. Conservation indicates that a sequence has been maintained by natural selection.

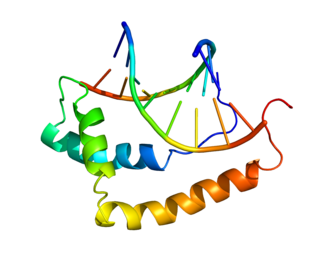
T-box transcription factor T, also known as Brachyury protein, is encoded for in humans by the TBXT gene. Brachyury functions as a transcription factor within the T-box family of genes. Brachyury homologs have been found in all bilaterian animals that have been screened, as well as the freshwater cnidarian Hydra.

Serine/threonine kinase 11 (STK11) also known as liver kinase B1 (LKB1) or renal carcinoma antigen NY-REN-19 is a protein kinase that in humans is encoded by the STK11 gene.
Transcription factor SOX-9 is a protein that in humans is encoded by the SOX9 gene.

Long non-coding RNAs are a type of RNA, generally defined as transcripts more than 200 nucleotides that are not translated into protein. This arbitrary limit distinguishes long ncRNAs from small non-coding RNAs, such as microRNAs (miRNAs), small interfering RNAs (siRNAs), Piwi-interacting RNAs (piRNAs), small nucleolar RNAs (snoRNAs), and other short RNAs. Given that some lncRNAs have been reported to have the potential to encode small proteins or micro-peptides, the latest definition of lncRNA is a class of RNA molecules of over 200 nucleotides that have no or limited coding capacity. Long intervening/intergenic noncoding RNAs (lincRNAs) are sequences of lncRNA which do not overlap protein-coding genes.

HOTAIR is a human gene located between HOXC11 and HOXC12 on chromosome 12. It is the first example of an RNA expressed on one chromosome that has been found to influence transcription of HOXD cluster posterior genes located on chromosome 2. The sequence and function of HOTAIR is different in human and mouse. Sequence analysis of HOTAIR revealed that it exists in mammals, has poorly conserved sequences and considerably conserved structures, and has evolved faster than nearby HoxC genes. A subsequent study identified HOTAIR has 32 nucleotide long conserved noncoding element (CNE) that has a paralogous copy in HOXD cluster region, suggesting that the HOTAIR conserved sequences predates whole genome duplication events at the root of vertebrate. While the conserved sequence paralogous with HOXD cluster is 32 nucleotide long, the HOTAIR sequence conserved from human to fish is about 200 nucleotide long and is marked by active enhancer features.
UCbase is a database of ultraconserved sequences that were first described by Bejerano, G. et al. in 2004. They are highly conserved genome regions that share 100% identity among human, mouse and rat. UCRs are 481 sequences longer than 200 bases. They are frequently located at genomic regions involved in cancer, differentially expressed in human leukemias and carcinomas and in some instances regulated by microRNAs. The first release of UCbase was published by Taccioli, C. et al. in 2009. Recent updates include new annotation based on hg19 Human genome, information about disorders related to the chromosome coordinates using the SNOMED CT classification, a query tool to search for SNPs, and a new text box to directly interrogate the database using a MySQL interface. Moreover, a sequence comparison tool allows the researchers to match selected sequences against ultraconserved elements located in genomic regions involved in specific disorders. To facilitate the interactive, visual interpretation of UCR chromosomal coordinates, the authors have implemented the graph visualization feature of UCbase creating a link to the UCSC Genome Browser. UCbase 2.0 does not provide microRNAs (miRNAs) information anymore focusing only on UCRs. The official release of UCbase 2.0 was published in 2014.
A conserved non-coding sequence (CNS) is a DNA sequence of noncoding DNA that is evolutionarily conserved. These sequences are of interest for their potential to regulate gene production.

CDKN2B-AS, also known as ANRIL is a long non-coding RNA consisting of 19 exons, spanning 126.3kb in the genome, and its spliced product is a 3834bp RNA. It is located within the p15/CDKN2B-p16/CDKN2A-p14/ARF gene cluster, in the antisense direction. Single nucleotide polymorphisms (SNPs) which alter the expression of CDKN2B-AS are associated with human healthy life expectancy, as well as with multiple diseases, including coronary artery disease, diabetes and many cancers. It binds to chromobox 7 (CBX7) within the polycomb repressive complex 1 and to SUZ12, a component of polycomb repression complex 2 and through these interactions is involved in transcriptional repression.
TUC338 is an ultra-conserved element which is transcribed to give a non-coding RNA. The TUC338 gene was first identified as uc.338, along with 480 other ultra-conserved elements in the human genome. Expression of this RNA gene has been found to dramatically increase in hepatocellular carcinoma (HCC) cells.

Long interspersed nuclear elements (LINEs) are a group of non-LTR retrotransposons that are widespread in the genome of many eukaryotes. LINEs contain an internal Pol II promoter to initiate transcription into mRNA, and encode one or two proteins, ORF1 and ORF2. The functional domains present within ORF1 vary greatly among LINEs, but often exhibit RNA/DNA binding activity. ORF2 is essential to successful retrotransposition, and encodes a protein with both reverse transcriptase and endonuclease activity.

LINE1 is a family of related class I transposable elements in the DNA of some organisms, classified with the long interspersed elements (LINEs). L1 transposons comprise approximately 17% of the human genome. These active L1s can interrupt the genome through insertions, deletions, rearrangements, and copy number variations. L1 activity has contributed to the instability and evolution of genomes and is tightly regulated in the germline by DNA methylation, histone modifications, and piRNA. L1s can further impact genome variation through mispairing and unequal crossing over during meiosis due to its repetitive DNA sequences.

Short interspersed nuclear elements (SINEs) are non-autonomous, non-coding transposable elements (TEs) that are about 100 to 700 base pairs in length. They are a class of retrotransposons, DNA elements that amplify themselves throughout eukaryotic genomes, often through RNA intermediates. SINEs compose about 13% of the mammalian genome.
Generally, in progression to cancer, hundreds of genes are silenced or activated. Although silencing of some genes in cancers occurs by mutation, a large proportion of carcinogenic gene silencing is a result of altered DNA methylation. DNA methylation causing silencing in cancer typically occurs at multiple CpG sites in the CpG islands that are present in the promoters of protein coding genes.

Solute carrier family 39 member 12 is a protein that in humans is encoded by the SLC39A12 gene.
References
- 1 2 3 4 5 6 Reneker J, Lyons E, Conant GC, Pires JC, Freeling M, Shyu CR, Korkin D (May 2012). "Long identical multispecies elements in plant and animal genomes". Proceedings of the National Academy of Sciences of the United States of America. 109 (19): E1183–E1191. doi: 10.1073/pnas.1121356109 . PMC 3358895 . PMID 22496592.
- 1 2 3 4 5 6 7 8 9 10 11 12 13 Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D (May 2004). "Ultraconserved elements in the human genome". Science. 304 (5675): 1321–1325. Bibcode:2004Sci...304.1321B. CiteSeerX 10.1.1.380.9305 . doi:10.1126/science.1098119. PMID 15131266. S2CID 2790337.
- 1 2 3 Pereira Zambalde E, Mathias C, Rodrigues AC, de Souza Fonseca Ribeiro EM, Fiori Gradia D, Calin GA, Carvalho de Oliveira J (March 2020). "Highlighting transcribed ultraconserved regions in human diseases". Wiley Interdisciplinary Reviews. RNA. 11 (2): e1567. doi:10.1002/wrna.1567. PMID 31489780. S2CID 201844414.
- ↑ Taccioli C, Fabbri E, Visone R, Volinia S, Calin GA, Fong LY, et al. (January 2009). "UCbase & miRfunc: a database of ultraconserved sequences and microRNA function". Nucleic Acids Research. 37 (Database issue): D41–D48. doi:10.1093/nar/gkn702. PMC 2686429 . PMID 18945703.
- 1 2 Katzman S, Kern AD, Bejerano G, Fewell G, Fulton L, Wilson RK, et al. (August 2007). "Human genome ultraconserved elements are ultraselected". Science. 317 (5840): 915. Bibcode:2007Sci...317..915K. doi:10.1126/science.1142430. PMID 17702936. S2CID 35322654.
- 1 2 3 4 5 6 Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, et al. (September 2007). "Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas". Cancer Cell. 12 (3): 215–229. doi: 10.1016/j.ccr.2007.07.027 . PMID 17785203.
- ↑ Sathirapongsasuti JF, Sathira N, Suzuki Y, Huttenhower C, Sugano S (March 2011). "Ultraconserved cDNA segments in the human transcriptome exhibit resistance to folding and implicate function in translation and alternative splicing". Nucleic Acids Research. 39 (6): 1967–1979. doi:10.1093/nar/gkq949. PMC 3064809 . PMID 21062826.
- 1 2 3 Habic A, Mattick JS, Calin GA, Krese R, Konc J, Kunej T (November 2019). "Genetic Variations of Ultraconserved Elements in the Human Genome". Omics. 23 (11): 549–559. doi:10.1089/omi.2019.0156. PMC 6857462 . PMID 31689173.
- 1 2 Ahituv N, Zhu Y, Visel A, Holt A, Afzal V, Pennacchio LA, Rubin EM (September 2007). "Deletion of ultraconserved elements yields viable mice". PLOS Biology. 5 (9): e234. doi: 10.1371/journal.pbio.0050234 . PMC 1964772 . PMID 17803355.
- ↑ Snetkova V, Ypsilanti AR, Akiyama JA, Mannion BJ, Plajzer-Frick I, Novak CS, et al. (April 2021). "Ultraconserved enhancer function does not require perfect sequence conservation". Nature Genetics. 53 (4): 521–528. doi:10.1038/s41588-021-00812-3. PMC 8038972 . PMID 33782603.
- 1 2 3 4 Fedorova L, Mulyar OA, Lim J, Fedorov A (November 2022). "Nucleotide Composition of Ultra-Conserved Elements Shows Excess of GpC and Depletion of GG and CC Dinucleotides". Genes. 13 (11): 2053. doi: 10.3390/genes13112053 . PMC 9690913 . PMID 36360290.
- 1 2 Saygin D, Tabib T, Bittar HE, Valenzi E, Sembrat J, Chan SY, et al. (January 2005). "Transcriptional profiling of lung cell populations in idiopathic pulmonary arterial hypertension". Pulmonary Circulation. 10 (1): e19. doi: 10.1371/journal.pbio.0030019 . PMC 544543 . PMID 32166015.

- ↑ Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, et al. (January 2005). "Highly conserved non-coding sequences are associated with vertebrate development". PLOS Biology. 3 (1): e7. doi: 10.1371/journal.pbio.0030007 . PMC 526512 . PMID 15630479.

- ↑ Elizabeth Pennisi (2017) Mysterious unchanging DNA finds a purpose in life, Science 02 Jun 2017]
- ↑ Braconi C, Valeri N, Kogure T, Gasparini P, Huang N, Nuovo GJ, et al. (January 2011). "Expression and functional role of a transcribed noncoding RNA with an ultraconserved element in hepatocellular carcinoma". Proceedings of the National Academy of Sciences of the United States of America. 108 (2): 786–791. Bibcode:2011PNAS..108..786B. doi: 10.1073/pnas.1011098108 . PMC 3021052 . PMID 21187392.
- ↑ McCole RB, Fonseka CY, Koren A, Wu CT (October 2014). "Abnormal dosage of ultraconserved elements is highly disfavored in healthy cells but not cancer cells". PLOS Genetics. 10 (10): e1004646. doi: 10.1371/journal.pgen.1004646 . PMC 4207606 . PMID 25340765.
- ↑ Derti A, Roth FP, Church GM, Wu CT (October 2006). "Mammalian ultraconserved elements are strongly depleted among segmental duplications and copy number variants". Nature Genetics. 38 (10): 1216–1220. doi:10.1038/ng1888. PMID 16998490. S2CID 10671674.
- ↑ Chiang CW, Derti A, Schwartz D, Chou MF, Hirschhorn JN, Wu CT (December 2008). "Ultraconserved elements: analyses of dosage sensitivity, motifs and boundaries". Genetics. 180 (4): 2277–2293. doi:10.1534/genetics.108.096537. PMC 2600958 . PMID 18957701.
- ↑ De Grassi A, Segala C, Iannelli F, Volorio S, Bertario L, Radice P, et al. (January 2010). Hastie N (ed.). "Ultradeep sequencing of a human ultraconserved region reveals somatic and constitutional genomic instability". PLOS Biology. 8 (1): e1000275. doi: 10.1371/journal.pbio.1000275 . PMC 2794366 . PMID 20052272.
- 1 2 3 Sekino Y, Sakamoto N, Goto K, Honma R, Shigematsu Y, Sentani K, et al. (November 2017). "Transcribed ultraconserved region Uc.63+ promotes resistance to docetaxel through regulation of androgen receptor signaling in prostate cancer". Oncotarget. 8 (55): 94259–94270. doi:10.18632/oncotarget.21688. PMC 5706872 . PMID 29212226.
- 1 2 Goto K, Ishikawa S, Honma R, Tanimoto K, Sakamoto N, Sentani K, et al. (July 2016). "The transcribed-ultraconserved regions in prostate and gastric cancer: DNA hypermethylation and microRNA-associated regulation" (PDF). Oncogene. 35 (27): 3598–3606. doi:10.1038/onc.2015.445. PMID 26640143. S2CID 8494774.
- ↑ Pang W, Su J, Wang Y, Feng H, Dai X, Yuan Y, et al. (October 2015). "Pancreatic cancer-secreted miR-155 implicates in the conversion from normal fibroblasts to cancer-associated fibroblasts". Cancer Science. 106 (10): 1362–1369. doi:10.1111/cas.12747. PMC 4638007 . PMID 26195069.
- ↑ Liz J, Portela A, Soler M, Gómez A, Ling H, Michlewski G, et al. (July 2014). "Regulation of pri-miRNA processing by a long noncoding RNA transcribed from an ultraconserved region". Molecular Cell. 55 (1): 138–147. doi: 10.1016/j.molcel.2014.05.005 . PMID 24910097.
- ↑ Guo J, Fang W, Sun L, Lu Y, Dou L, Huang X, et al. (February 2018). "Ultraconserved element uc.372 drives hepatic lipid accumulation by suppressing miR-195/miR4668 maturation". Nature Communications. 9 (1): 612. Bibcode:2018NatCo...9..612G. doi:10.1038/s41467-018-03072-8. PMC 5807361 . PMID 29426937.
- ↑ Xiao L, Wu J, Wang JY, Chung HK, Kalakonda S, Rao JN, et al. (February 2018). "Long Noncoding RNA uc.173 Promotes Renewal of the Intestinal Mucosa by Inducing Degradation of MicroRNA 195". Gastroenterology. 154 (3): 599–611. doi:10.1053/j.gastro.2017.10.009. PMC 5811324 . PMID 29042220.
- ↑ Wojcik SE, Rossi S, Shimizu M, Nicoloso MS, Cimmino A, Alder H, et al. (February 2010). "Non-codingRNA sequence variations in human chronic lymphocytic leukemia and colorectal cancer". Carcinogenesis. 31 (2): 208–215. doi:10.1093/carcin/bgp209. PMC 2812567 . PMID 19926640.
- ↑ Nan A, Zhou X, Chen L, Liu M, Zhang N, Zhang L, et al. (January 2016). "A transcribed ultraconserved noncoding RNA, Uc.173, is a key molecule for the inhibition of lead-induced neuronal apoptosis". Oncotarget. 7 (1): 112–124. doi:10.18632/oncotarget.6590. PMC 4807986 . PMID 26683706.
- ↑ Wang JY, Cui YH, Xiao L, Chung HK, Zhang Y, Rao JN, et al. (July 2018). "Regulation of Intestinal Epithelial Barrier Function by Long Noncoding RNA uc.173 through Interaction with MicroRNA 29b". Molecular and Cellular Biology. 38 (13): e00010–18. doi:10.1128/MCB.00010-18. PMC 6002690 . PMID 29632078.
- ↑ Vannini I, Wise PM, Challagundla KB, Plousiou M, Raffini M, Bandini E, et al. (November 2017). "Transcribed ultraconserved region 339 promotes carcinogenesis by modulating tumor suppressor microRNAs". Nature Communications. 8 (1): 1801. Bibcode:2017NatCo...8.1801V. doi:10.1038/s41467-017-01562-9. PMC 5703849 . PMID 29180617.
- ↑ Olivieri M, Ferro M, Terreri S, Durso M, Romanelli A, Avitabile C, et al. (April 2016). "Long non-coding RNA containing ultraconserved genomic region 8 promotes bladder cancer tumorigenesis". Oncotarget. 7 (15): 20636–20654. doi:10.18632/oncotarget.7833. PMC 4991481 . PMID 26943042.
- ↑ Kottorou AE, Antonacopoulou AG, Dimitrakopoulos FD, Diamantopoulou G, Sirinian C, Kalofonou M, et al. (April 2018). "Deregulation of methylation of transcribed-ultra conserved regions in colorectal cancer and their value for detection of adenomas and adenocarcinomas". Oncotarget. 9 (30): 21411–21428. doi:10.18632/oncotarget.25115. PMC 5940382 . PMID 29765549.
- ↑ Kajita K, Kuwano Y, Satake Y, Kano S, Kurokawa K, Akaike Y, et al. (April 2016). "Ultraconserved region-containing Transformer 2β4 controls senescence of colon cancer cells". Oncogenesis. 5 (4): e213. doi:10.1038/oncsis.2016.18. PMC 4848834 . PMID 27043659.
- ↑ Terreri S, Durso M, Colonna V, Romanelli A, Terracciano D, Ferro M, et al. (December 2016). "New Cross-Talk Layer between Ultraconserved Non-Coding RNAs, MicroRNAs and Polycomb Protein YY1 in Bladder Cancer". Genes. 7 (12): 127. doi: 10.3390/genes7120127 . PMC 5192503 . PMID 27983635.
- ↑ Cronin S, Berger S, Ding J, Schymick JC, Washecka N, Hernandez DG, et al. (March 2008). "A genome-wide association study of sporadic ALS in a homogenous Irish population". Human Molecular Genetics. 17 (5): 768–774. doi: 10.1093/hmg/ddm361 . PMID 18057069.
- ↑ Hansen MF, Neckmann U, Lavik LA, Vold T, Gilde B, Toft RK, Sjursen W (March 2014). "A massive parallel sequencing workflow for diagnostic genetic testing of mismatch repair genes". Molecular Genetics & Genomic Medicine. 2 (2): 186–200. doi:10.1002/mgg3.62. PMC 3960061 . PMID 24689082.
- ↑ Chiang CW, Liu CT, Lettre G, Lange LA, Jorgensen NW, Keating BJ, et al. (September 2012). "Ultraconserved elements in the human genome: association and transmission analyses of highly constrained single-nucleotide polymorphisms". Genetics. 192 (1): 253–266. doi:10.1534/genetics.112.141945. PMC 3430540 . PMID 22714408.
- ↑ Khor CC, Miyake M, Chen LJ, Shi Y, Barathi VA, Qiao F, et al. (December 2013). "Genome-wide association study identifies ZFHX1B as a susceptibility locus for severe myopia". Human Molecular Genetics. 22 (25): 5288–5294. doi: 10.1093/hmg/ddt385 . PMID 23933737.
- 1 2 Dobbs MB, Gurnett CA, Pierce B, Exner GU, Robarge J, Morcuende JA, et al. (March 2006). "HOXD10 M319K mutation in a family with isolated congenital vertical talus". Journal of Orthopaedic Research. 24 (3): 448–453. doi: 10.1002/jor.20052 . PMID 16450407. S2CID 28670628.
- ↑ Lu Y, Vitart V, Burdon KP, Khor CC, Bykhovskaya Y, Mirshahi A, et al. (February 2013). "Genome-wide association analyses identify multiple loci associated with central corneal thickness and keratoconus". Nature Genetics. 45 (2): 155–163. doi:10.1038/ng.2506. PMC 3720123 . PMID 23291589.
- 1 2 3 Bosch DG, Boonstra FN, Gonzaga-Jauregui C, Xu M, de Ligt J, Jhangiani S, et al. (February 2014). "NR2F1 mutations cause optic atrophy with intellectual disability". American Journal of Human Genetics. 94 (2): 303–309. doi:10.1016/j.ajhg.2014.01.002. PMC 3928641 . PMID 24462372.
- ↑ Soravia C, Sugg SL, Berk T, Mitri A, Cheng H, Gallinger S, et al. (January 1999). "Familial adenomatous polyposis-associated thyroid cancer: a clinical, pathological, and molecular genetics study". The American Journal of Pathology. 154 (1): 127–135. doi:10.1016/S0002-9440(10)65259-5. PMC 1853451 . PMID 9916927.
- ↑ Chang YS, Lin CY, Yang SF, Ho CM, Chang JG (2016-03-28). "Analysing the mutational status of adenomatous polyposis coli (APC) gene in breast cancer". Cancer Cell International. 16: 23. doi: 10.1186/s12935-016-0297-2 . PMC 4810512 . PMID 27028212.
- ↑ Fodde R, van der Luijt R, Wijnen J, Tops C, van der Klift H, van Leeuwen-Cornelisse I, et al. (August 1992). "Eight novel inactivating germ line mutations at the APC gene identified by denaturing gradient gel electrophoresis". Genomics. 13 (4): 1162–1168. doi:10.1016/0888-7543(92)90032-n. PMID 1324223.
- ↑ Curia MC, Esposito DL, Aceto G, Palmirotta R, Crognale S, Valanzano R, et al. (1998). "Transcript dosage effect in familial adenomatous polyposis: model offered by two kindreds with exon 9 APC gene mutations". Human Mutation. 11 (3): 197–201. doi:10.1002/(SICI)1098-1004(1998)11:3<197::AID-HUMU3>3.0.CO;2-F. PMID 9521420. S2CID 7241178.
- ↑ Johnson JO, Pioro EP, Boehringer A, Chia R, Feit H, Renton AE, et al. (May 2014). "Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis". Nature Neuroscience. 17 (5): 664–666. doi:10.1038/nn.3688. PMC 4000579 . PMID 24686783.
- ↑ Au PY, You J, Caluseriu O, Schwartzentruber J, Majewski J, Bernier FP, et al. (October 2015). "GeneMatcher aids in the identification of a new malformation syndrome with intellectual disability, unique facial dysmorphisms, and skeletal and connective tissue abnormalities caused by de novo variants in HNRNPK". Human Mutation. 36 (10): 1009–1014. doi:10.1002/humu.22837. PMC 4589226 . PMID 26173930.
- ↑ Mallery DL, Tanganelli B, Colella S, Steingrimsdottir H, van Gool AJ, Troelstra C, et al. (January 1998). "Molecular analysis of mutations in the CSB (ERCC6) gene in patients with Cockayne syndrome". American Journal of Human Genetics. 62 (1): 77–85. doi:10.1086/301686. PMC 1376810 . PMID 9443879.
- ↑ Schimmenti LA, Shim HH, Wirtschafter JD, Panzarino VA, Kashtan CE, Kirkpatrick SJ, et al. (1999). "Homonucleotide expansion and contraction mutations of PAX2 and inclusion of Chiari 1 malformation as part of renal-coloboma syndrome". Human Mutation. 14 (5): 369–376. doi: 10.1002/(SICI)1098-1004(199911)14:5<369::AID-HUMU2>3.0.CO;2-E . PMID 10533062. S2CID 25564812.
- ↑ Amiel J, Audollent S, Joly D, Dureau P, Salomon R, Tellier AL, et al. (November 2000). "PAX2 mutations in renal-coloboma syndrome: mutational hotspot and germline mosaicism". European Journal of Human Genetics. 8 (11): 820–826. doi: 10.1038/sj.ejhg.5200539 . PMID 11093271. S2CID 30359554.
- ↑ Schimmenti LA, Cunliffe HE, McNoe LA, Ward TA, French MC, Shim HH, et al. (April 1997). "Further delineation of renal-coloboma syndrome in patients with extreme variability of phenotype and identical PAX2 mutations". American Journal of Human Genetics. 60 (4): 869–878. PMC 1712484 . PMID 9106533.
- ↑ Barua M, Stellacci E, Stella L, Weins A, Genovese G, Muto V, et al. (September 2014). "Mutations in PAX2 associate with adult-onset FSGS". Journal of the American Society of Nephrology. 25 (9): 1942–1953. doi:10.1681/ASN.2013070686. PMC 4147972 . PMID 24676634.
- ↑ Chettier R, Nelson L, Ogilvie JW, Albertsen HM, Ward K (2015-02-12). Fang S (ed.). "Haplotypes at LBX1 have distinct inheritance patterns with opposite effects in adolescent idiopathic scoliosis". PLOS ONE. 10 (2): e0117708. Bibcode:2015PLoSO..1017708C. doi: 10.1371/journal.pone.0117708 . PMC 4326419 . PMID 25675428.
- ↑ Gao W, Peng Y, Liang G, Liang A, Ye W, Zhang L, et al. (2013-01-04). "Association between common variants near LBX1 and adolescent idiopathic scoliosis replicated in the Chinese Han population". PLOS ONE. 8 (1): e53234. Bibcode:2013PLoSO...853234G. doi: 10.1371/journal.pone.0053234 . PMC 3537668 . PMID 23308168.
- ↑ Grauers A, Wang J, Einarsdottir E, Simony A, Danielsson A, Åkesson K, et al. (October 2015). "Candidate gene analysis and exome sequencing confirm LBX1 as a susceptibility gene for idiopathic scoliosis". The Spine Journal. 15 (10): 2239–2246. doi:10.1016/j.spinee.2015.05.013. hdl: 10616/44765 . PMID 25987191.
- ↑ Jiang H, Qiu X, Dai J, Yan H, Zhu Z, Qian B, Qiu Y (February 2013). "Association of rs11190870 near LBX1 with adolescent idiopathic scoliosis susceptibility in a Han Chinese population". European Spine Journal. 22 (2): 282–286. doi:10.1007/s00586-012-2532-4. PMC 3555620 . PMID 23096252.
- ↑ Londono D, Kou I, Johnson TA, Sharma S, Ogura Y, Tsunoda T, et al. (June 2014). "A meta-analysis identifies adolescent idiopathic scoliosis association with LBX1 locus in multiple ethnic groups". Journal of Medical Genetics. 51 (6): 401–406. doi:10.1136/jmedgenet-2013-102067. PMID 24721834. S2CID 23646905.
- ↑ Miyake A, Kou I, Takahashi Y, Johnson TA, Ogura Y, Dai J, et al. (2013-09-04). "Identification of a susceptibility locus for severe adolescent idiopathic scoliosis on chromosome 17q24.3". PLOS ONE. 8 (9): e72802. Bibcode:2013PLoSO...872802M. doi: 10.1371/journal.pone.0072802 . PMC 3762929 . PMID 24023777.
- ↑ Jiang Y, Ben Q, Shen H, Lu W, Zhang Y, Zhu J (November 2011). "Diabetes mellitus and incidence and mortality of colorectal cancer: a systematic review and meta-analysis of cohort studies". European Journal of Epidemiology. 26 (11): 863–876. doi:10.1007/s10654-011-9617-y. PMID 21938478. S2CID 99605.
- ↑ Takahashi Y, Kou I, Takahashi A, Johnson TA, Kono K, Kawakami N, et al. (October 2011). "A genome-wide association study identifies common variants near LBX1 associated with adolescent idiopathic scoliosis". Nature Genetics. 43 (12): 1237–1240. doi:10.1038/ng.974. PMID 22019779. S2CID 7533298.
- ↑ Buchert R, Tawamie H, Smith C, Uebe S, Innes AM, Al Hallak B, et al. (November 2014). "A peroxisomal disorder of severe intellectual disability, epilepsy, and cataracts due to fatty acyl-CoA reductase 1 deficiency". American Journal of Human Genetics. 95 (5): 602–610. doi:10.1016/j.ajhg.2014.10.003. PMC 4225589 . PMID 25439727.
- ↑ O'Donnell PH, Stark AL, Gamazon ER, Wheeler HE, McIlwee BE, Gorsic L, et al. (August 2012). "Identification of novel germline polymorphisms governing capecitabine sensitivity". Cancer. 118 (16): 4063–4073. doi:10.1002/cncr.26737. PMC 3413892 . PMID 22864933.
- ↑ De R, Verma SS, Drenos F, Holzinger ER, Holmes MV, Hall MA, et al. (June 2015). "Identifying gene-gene interactions that are highly associated with Body Mass Index using Quantitative Multifactor Dimensionality Reduction (QMDR)". BioData Mining. 8 (1): 41. doi: 10.1186/s13040-015-0074-0 . PMC 4678717 . PMID 26674805.
- ↑ Guo Y, Lanktree MB, Taylor KC, Hakonarson H, Lange LA, Keating BJ (January 2013). "Gene-centric meta-analyses of 108 912 individuals confirm known body mass index loci and reveal three novel signals". Human Molecular Genetics. 22 (1): 184–201. doi:10.1093/hmg/dds396. PMC 3522401 . PMID 23001569.
- ↑ Hromatka BS, Tung JY, Kiefer AK, Do CB, Hinds DA, Eriksson N (May 2015). "Genetic variants associated with motion sickness point to roles for inner ear development, neurological processes and glucose homeostasis". Human Molecular Genetics. 24 (9): 2700–2708. doi:10.1093/hmg/ddv028. PMC 4383869 . PMID 25628336.
- ↑ Al Turki S, Manickaraj AK, Mercer CL, Gerety SS, Hitz MP, Lindsay S, et al. (April 2014). "Rare variants in NR2F2 cause congenital heart defects in humans". American Journal of Human Genetics. 94 (4): 574–585. doi:10.1016/j.ajhg.2014.03.007. PMC 3980509 . PMID 24702954.
- ↑ Hofstra RM, Mulder IM, Vossen R, de Koning-Gans PA, Kraak M, Ginjaar IB, et al. (January 2004). "DGGE-based whole-gene mutation scanning of the dystrophin gene in Duchenne and Becker muscular dystrophy patients". Human Mutation. 23 (1): 57–66. doi: 10.1002/humu.10283 . PMID 14695533. S2CID 36020079.
- ↑ Roberts RG, Bobrow M, Bentley DR (March 1992). "Point mutations in the dystrophin gene". Proceedings of the National Academy of Sciences of the United States of America. 89 (6): 2331–2335. Bibcode:1992PNAS...89.2331R. doi: 10.1073/pnas.89.6.2331 . PMC 48651 . PMID 1549596.
- ↑ Tuffery-Giraud S, Saquet C, Thorel D, Disset A, Rivier F, Malcolm S, Claustres M (December 2005). "Mutation spectrum leading to an attenuated phenotype in dystrophinopathies". European Journal of Human Genetics. 13 (12): 1254–1260. doi: 10.1038/sj.ejhg.5201478 . PMID 16077730. S2CID 22585201.
- ↑ Gorman MP, Golomb MR, Walsh LE, Hobson GM, Garbern JY, Kinkel RP, et al. (April 2007). "Steroid-responsive neurologic relapses in a child with a proteolipid protein-1 mutation". Neurology. 68 (16): 1305–1307. doi:10.1212/01.wnl.0000259522.49388.53. PMID 17438221. S2CID 45639125.
- ↑ Saugier-Veber P, Munnich A, Bonneau D, Rozet JM, Le Merrer M, Gil R, Boespflug-Tanguy O (March 1994). "X-linked spastic paraplegia and Pelizaeus-Merzbacher disease are allelic disorders at the proteolipid protein locus". Nature Genetics. 6 (3): 257–262. doi:10.1038/ng0394-257. PMID 8012387. S2CID 13607673.
- ↑ Hodes ME, Blank CA, Pratt VM, Morales J, Napier J, Dlouhy SR (March 1997). "Nonsense mutation in exon 3 of the proteolipid protein gene (PLP) in a family with an unusual form of Pelizaeus-Merzbacher disease". American Journal of Medical Genetics. 69 (2): 121–125. doi:10.1002/(SICI)1096-8628(19970317)69:2<121::AID-AJMG2>3.0.CO;2-S. PMID 9056547.
- ↑ Wu Y, Arai AC, Rumbaugh G, Srivastava AK, Turner G, Hayashi T, et al. (November 2007). "Mutations in ionotropic AMPA receptor 3 alter channel properties and are associated with moderate cognitive impairment in humans". Proceedings of the National Academy of Sciences of the United States of America. 104 (46): 18163–18168. Bibcode:2007PNAS..10418163W. doi: 10.1073/pnas.0708699104 . PMC 2084314 . PMID 17989220.
- ↑ Gueneau L, Bertrand AT, Jais JP, Salih MA, Stojkovic T, Wehnert M, et al. (September 2009). "Mutations of the FHL1 gene cause Emery-Dreifuss muscular dystrophy". American Journal of Human Genetics. 85 (3): 338–353. doi:10.1016/j.ajhg.2009.07.015. PMC 2771595 . PMID 19716112.
- ↑ Knoblauch H, Geier C, Adams S, Budde B, Rudolph A, Zacharias U, et al. (January 2010). "Contractures and hypertrophic cardiomyopathy in a novel FHL1 mutation". Annals of Neurology. 67 (1): 136–140. doi:10.1002/ana.21839. PMID 20186852. S2CID 30441775.
- 1 2 Schoser B, Goebel HH, Janisch I, Quasthoff S, Rother J, Bergmann M, et al. (August 2009). "Consequences of mutations within the C terminus of the FHL1 gene". Neurology. 73 (7): 543–551. doi:10.1212/WNL.0b013e3181b2a4b3. PMID 19687455. S2CID 13107330.
- 1 2 Windpassinger C, Schoser B, Straub V, Hochmeister S, Noor A, Lohberger B, et al. (January 2008). "An X-linked myopathy with postural muscle atrophy and generalized hypertrophy, termed XMPMA, is caused by mutations in FHL1". American Journal of Human Genetics. 82 (1): 88–99. doi:10.1016/j.ajhg.2007.09.004. PMC 2253986 . PMID 18179888.
- ↑ Zahorakova D, Rosipal R, Hadac J, Zumrova A, Bzduch V, Misovicova N, et al. (2007). "Mutation analysis of the MECP2 gene in patients of Slavic origin with Rett syndrome: novel mutations and polymorphisms". Journal of Human Genetics. 52 (4): 342–348. doi: 10.1007/s10038-007-0121-x . PMID 17387578. S2CID 7962500.