Related Research Articles
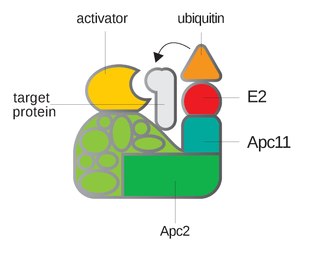
Anaphase-promoting complex is an E3 ubiquitin ligase that marks target cell cycle proteins for degradation by the 26S proteasome. The APC/C is a large complex of 11–13 subunit proteins, including a cullin (Apc2) and RING (Apc11) subunit much like SCF. Other parts of the APC/C have unknown functions but are highly conserved.

The G0 phase describes a cellular state outside of the replicative cell cycle. Classically, cells were thought to enter G0 primarily due to environmental factors, like nutrient deprivation, that limited the resources necessary for proliferation. Thus it was thought of as a resting phase. G0 is now known to take different forms and occur for multiple reasons. For example, most adult neuronal cells, among the most metabolically active cells in the body, are fully differentiated and reside in a terminal G0 phase. Neurons reside in this state, not because of stochastic or limited nutrient supply, but as a part of their developmental program.

Cyclin-dependent kinases (CDKs) are the families of protein kinases first discovered for their role in regulating the cell cycle. They are also involved in regulating transcription, mRNA processing, and the differentiation of nerve cells. They are present in all known eukaryotes, and their regulatory function in the cell cycle has been evolutionarily conserved. In fact, yeast cells can proliferate normally when their CDK gene has been replaced with the homologous human gene. CDKs are relatively small proteins, with molecular weights ranging from 34 to 40 kDa, and contain little more than the kinase domain. By definition, a CDK binds a regulatory protein called a cyclin. Without cyclin, CDK has little kinase activity; only the cyclin-CDK complex is an active kinase but its activity can be typically further modulated by phosphorylation and other binding proteins, like p27. CDKs phosphorylate their substrates on serines and threonines, so they are serine-threonine kinases. The consensus sequence for the phosphorylation site in the amino acid sequence of a CDK substrate is [S/T*]PX[K/R], where S/T* is the phosphorylated serine or threonine, P is proline, X is any amino acid, K is lysine, and R is arginine.
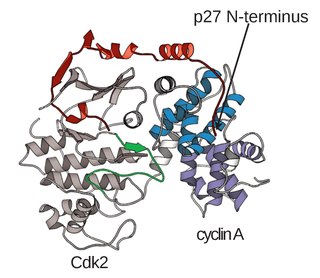
A cyclin-dependent kinase complex is a protein complex formed by the association of an inactive catalytic subunit of a protein kinase, cyclin-dependent kinase (CDK), with a regulatory subunit, cyclin. Once cyclin-dependent kinases bind to cyclin, the formed complex is in an activated state. Substrate specificity of the activated complex is mainly established by the associated cyclin within the complex. Activity of CDKCs is controlled by phosphorylation of target proteins, as well as binding of inhibitory proteins.
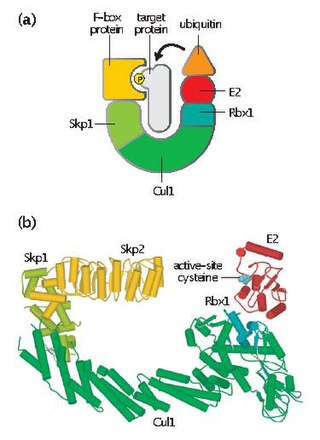
Skp, Cullin, F-box containing complex is a multi-protein E3 ubiquitin ligase complex that catalyzes the ubiquitination of proteins destined for 26S proteasomal degradation. Along with the anaphase-promoting complex, SCF has important roles in the ubiquitination of proteins involved in the cell cycle. The SCF complex also marks various other cellular proteins for destruction.

Cyclin D is a member of the cyclin protein family that is involved in regulating cell cycle progression. The synthesis of cyclin D is initiated during G1 and drives the G1/S phase transition. Cyclin D protein is anywhere from 155 to 477 amino acids in length.

Cyclin-dependent kinase 1 also known as CDK1 or cell division cycle protein 2 homolog is a highly conserved protein that functions as a serine/threonine protein kinase, and is a key player in cell cycle regulation. It has been highly studied in the budding yeast S. cerevisiae, and the fission yeast S. pombe, where it is encoded by genes cdc28 and cdc2, respectively. With its cyclin partners, Cdk1 forms complexes that phosphorylate a variety of target substrates ; phosphorylation of these proteins leads to cell cycle progression.
Polo-like kinases (Plks) are regulatory serine/threonin kinases of the cell cycle involved in mitotic entry, mitotic exit, spindle formation, cytokinesis, and meiosis. Only one Plk is found in the genomes of the fly Drosophila melanogaster (Polo), budding yeast (Cdc5) and fission yeast (Plo1). Vertebrates and other animals, however, have many Plk family members including Plk1, Plk2/Snk, Plk3/Prk/FnK, Plk4/Sak and Plk5. Of the vertebrate Plk family members, the mammalian Plk1 has been most extensively studied. During mitosis and cytokinesis, Plks associate with several structures including the centrosome, kinetochores, and the central spindle.
G1/S-specific cyclin Cln3 is a protein that is encoded by the CLN3 gene. The Cln3 protein is a budding yeast G1 cyclin that controls the timing of Start, the point of commitment to a mitotic cell cycle. It is an upstream regulator of the other G1 cyclins, and it is thought to be the key regulator linking cell growth to cell cycle progression. It is a 65 kD, unstable protein; like other cyclins, it functions by binding and activating cyclin-dependent kinase (CDK).
Sic1, a protein, is a stoichiometric inhibitor of Cdk1-Clb complexes in the budding yeast Saccharomyces cerevisiae. Because B-type cyclin-Cdk1 complexes are the drivers of S-phase initiation, Sic1 prevents premature S-phase entry. Multisite phosphorylation of Sic1 is thought to time Sic1 ubiquitination and destruction, and by extension, the timing of S-phase entry.

Wee1 is a nuclear kinase belonging to the Ser/Thr family of protein kinases in the fission yeast Schizosaccharomyces pombe. Wee1 has a molecular mass of 96 kDa and is a key regulator of cell cycle progression. It influences cell size by inhibiting the entry into mitosis, through inhibiting Cdk1. Wee1 has homologues in many other organisms, including mammals.
A series of biochemical switches control transitions between and within the various phases of the cell cycle. The cell cycle is a series of complex, ordered, sequential events that control how a single cell divides into two cells, and involves several different phases. The phases include the G1 and G2 phases, DNA replication or S phase, and the actual process of cell division, mitosis or M phase. During the M phase, the chromosomes separate and cytokinesis occurs.
The Start checkpoint is a major cell cycle checkpoint in yeast. The Start checkpoint ensures irreversible cell-cycle entry even if conditions later become unfavorable. The physiological factors that control passage through the Start checkpoint include external nutrient concentrations, presence of mating factor/ pheromone, forms of stress, and size control.

Cdc4 is a substrate recognition component of the SCF ubiquitin ligase complex, which acts as a mediator of ubiquitin transfer to target proteins, leading to their subsequent degradation via the ubiquitin-proteasome pathway. Cdc4 targets primarily cell cycle regulators for proteolysis. It serves the function of an adaptor that brings target molecules to the core SCF complex. Cdc4 was originally identified in the model organism Saccharomyces cerevisiae. CDC4 gene function is required at G1/S and G2/M transitions during mitosis and at various stages during meiosis.
Cdc14 and Cdc14 are a gene and its protein product respectively. Cdc14 is found in most of the eukaryotes. Cdc14 was defined by Hartwell in his famous screen for loci that control the cell cycle of Saccharomyces cerevisiae. Cdc14 was later shown to encode a protein phosphatase. Cdc14 is dual-specificity, which means it has serine/threonine and tyrosine-directed activity. A preference for serines next to proline is reported. Many early studies, especially in the budding yeast Saccharomyces cerevisiae, demonstrated that the protein plays a key role in regulating late mitotic processes. However, more recent work in a range of systems suggests that its cellular function is more complex.
Cln1, Cln2, and Cln3 are cyclin proteins expressed in the G1-phase of the cell cycle of budding yeast. Like other cyclins, they function by binding and activating cyclin-dependent kinase. They are responsible for initiating entry into a new mitotic cell cycle at Start. As described below, Cln3 is the primary regulator of this process during normal yeast growth, with the other two G1 cyclins performing their function upon induction by Cln3. However, Cln1 and Cln2 are also directly regulated by pathways sensing extracellular conditions, including the mating pheremone pathway.
Clb5 and Clb6 are B-type, S-phase cyclins in yeast that assist in cell cycle regulation. Clb5 and Clb6 bind and activate Cdk1, and high levels of these cyclins are required for entering S-phase. S-phase cyclin binding to Cdk1 directly stimulates DNA replication as well as progression to the next phase of the cell cycle.
Whi5 is a transcriptional regulator in the budding yeast cell cycle, notably in the G1 phase. It is an inhibitor of SBF, which is involved in the transcription of G1-specific genes. Cln3 promotes the disassociation of Whi5 from SBF, and its disassociation results in the transcription of genes needed to enter S phase.
Induced cell cycle arrest is the use of a chemical or genetic manipulation to artificially halt progression through the cell cycle. Cellular processes like genome duplication and cell division stop. It can be temporary or permanent. It is an artificial activation of naturally occurring cell cycle checkpoints, induced by exogenous stimuli controlled by an experimenter.
BCK2, also named CTR7, is an early cell cycle regulator expressed by the yeast Saccharomyces cerevisiae. It was first discovered in a screen for genes whose overexpression would suppress the phenotypes of PKC1 pathway mutations. Though its mechanism is currently unknown, it is believed to interact with Swi4 and Mcm1, both important transcriptional regulators of early cell cycle.
References
- ↑ "WHI3 - SGD". yeastgenome.org. Retrieved 19 May 2015.
- 1 2 Garí, E., Volpe, T., Wang, H., Gallego, C., Futcher, B., & Aldea, M. (2001). WHI3 binds the mRNA of the G1 cyclin CLN3 to modulate cell fate in budding yeast. Genes & development, 15(21), 2803-2808.
- ↑ Schneider B.L., Patton E.E., Lanker S., Mendenhall M.D., Wittenberg C., Futcher B., Tyers M.(1998) Yeast G1 cyclins are unstable in G1 phase. Nature 395:86–89.
- ↑ Colomina, N., Garí, E., Gallego, C., Herrero, E., & Aldea, M. (1999). G1 cyclins block the Ime1 pathway to make mitosis and meiosis incompatible in budding yeast. The EMBO Journal, 18(2), 320-329.
- 1 2 Wang, H., Garí, E., Verges, E., Gallego, C., & Aldea, M. (2004). Recruitment of Cdc28 by WHI3 restricts nuclear accumulation of the G1 cyclin–Cdk complex to late G1. The EMBO Journal, 23(1), 180-190.
- ↑ Caudron, F., & Barral, Y. (2013). A Super-Assembly of WHI3 Encodes Memory of Deceptive Encounters by Single Cells during Yeast Courtship. Cell, 155(6), 1244-1257.
- ↑ Nash, R. S., Volpe, T., & Futcher, B. (2001). Isolation and characterization of WHI3, a size-control gene of Saccharomyces cerevisiae. Genetics, 157(4), 1469-1480.