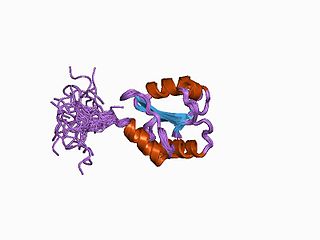
Protein disulfide isomerase, or PDI, is an enzyme in the endoplasmic reticulum (ER) in eukaryotes and the periplasm of bacteria that catalyzes the formation and breakage of disulfide bonds between cysteine residues within proteins as they fold. This allows proteins to quickly find the correct arrangement of disulfide bonds in their fully folded state, and therefore the enzyme acts to catalyze protein folding.
Matrix metalloproteinases (MMPs), also known as matrix metallopeptidases or matrixins, are metalloproteinases that are calcium-dependent zinc-containing endopeptidases; other family members are adamalysins, serralysins, and astacins. The MMPs belong to a larger family of proteases known as the metzincin superfamily.
In biology and biochemistry, protease inhibitors, or antiproteases, are molecules that inhibit the function of proteases. Many naturally occurring protease inhibitors are proteins.
Interleukins (ILs) are a group of cytokines that are expressed and secreted by white blood cells (leukocytes) as well as some other body cells. The human genome encodes more than 50 interleukins and related proteins.
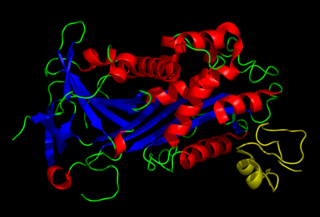
Vitronectin is a glycoprotein of the hemopexin family which is abundantly found in serum, the extracellular matrix and bone. In humans it is encoded by the VTN gene.

Protein disulfide-isomerase A3 (PDIA3), also known as glucose-regulated protein, 58-kD (GRP58), is an isomerase enzyme. This protein localizes to the endoplasmic reticulum (ER) and interacts with lectin chaperones calreticulin and calnexin (CNX) to modulate folding of newly synthesized glycoproteins. It is thought that complexes of lectins and this protein mediate protein folding by promoting formation of disulfide bonds in their glycoprotein substrates.

Matrilysin also known as matrix metalloproteinase-7 (MMP-7), pump-1 protease (PUMP-1), or uterine metalloproteinase is an enzyme in humans that is encoded by the MMP7 gene. The enzyme has also been known as matrin, putative metalloproteinase-1, matrix metalloproteinase pump 1, PUMP-1 proteinase, PUMP, metalloproteinase pump-1, putative metalloproteinase, MMP). Human MMP-7 has a molecular weight around 30 kDa.

Insulin-like growth factor-binding protein 3, also known as IGFBP-3, is a protein that in humans is encoded by the IGFBP3 gene. IGFBP-3 is one of six IGF binding proteins that have highly conserved structures and bind the insulin-like growth factors IGF-1 and IGF-2 with high affinity. IGFBP-7, sometimes included in this family, shares neither the conserved structural features nor the high IGF affinity. Instead, IGFBP-7 binds IGF1R, which blocks IGF-1 and IGF-2 binding, resulting in apoptosis.

Antileukoproteinase, also known as secretory leukocyte protease inhibitor (SLPI), is an enzyme that in humans is encoded by the SLPI gene. SLPI is a highly cationic single-chain protein with eight intramolecular disulfide bonds. It is found in large quantities in bronchial, cervical, and nasal mucosa, saliva, and seminal fluids. SLPI inhibits human leukocyte elastase, human cathepsin G, human trypsin, neutrophil elastase, and mast cell chymase. X-ray crystallography has shown that SLPI has two homologous domains of 53 and 54 amino acids, one of which exhibits anti-protease activity. The other domain is not known to have any function.

Elafin, also known as peptidase inhibitor 3 or skin-derived antileukoprotease (SKALP), is a protein that in humans is encoded by the PI3 gene.

WAP four-disulfide core domain protein 2 - also known as Human Epididymis Protein 4 (HE4) - is a protein that in humans is encoded by the WFDC2 gene.

WAP four-disulfide core domain protein 5 is a protein that in humans is encoded by the WFDC5 gene.

Eppin is a protein that in humans is encoded by the SPINLW1 gene.

WAP four-disulfide core domain protein 1 is a protein that in humans is encoded by the WFDC1 gene.
CUB domain is an evolutionarily conserved protein domain.
Somatomedin B is a serum factor of unknown function, is a small cysteine-rich peptide, derived proteolytically from the N-terminus of the cell-substrate adhesion protein vitronectin. Cys-rich somatomedin B-like domains are found in a number of proteins, including plasma-cell membrane glycoprotein and placental protein 11.
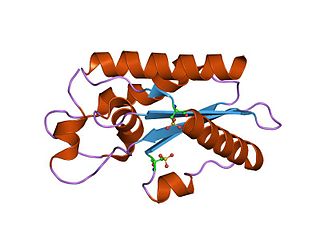
The CAP superfamily is a large superfamily of secreted proteins that are produced by a wide range of organisms, including prokaryotes and non-vertebrate eukaryotes.

In molecular biology, the trappin protein transglutaminase binding domain or cementoin is a protein domain found at the N-terminus of Whey Acidic Protein (WAP) domain-containing protease inhibitors such as trappin-2. This N-terminal domain enables it to become cross-linked to extracellular matrix proteins by transglutaminase. This domain contains several repeated motifs with the consensus sequence Gly-Gln-Asp-Pro-Val-Lys, and these together can anchor the whole molecule to extracellular matrix proteins, such as laminin, fibronectin, beta-crystallin, collagen IV, fibrinogen, and elastin, by transglutaminase-catalysed cross-links. The whole domain is rich in glutamine and lysine, thus allowing transglutaminase(s) to catalyse the formation of an intermolecular epsilon-(gamma-glutamyl)lysine isopeptide bond.
Cysteine-rich proteins are small proteins that contain a large number of cysteines. These cysteines either cross-link to form disulphide bonds, or bind metal ions by chelation, stabilising the protein's tertiary structure. CRPs include a highly conserved secretion peptide signal at the N-terminus and a cysteine-rich region at the C-terminus.

The ubiquitin carboxyl-terminal hydrolase 27, also known as deubiquitinating enzyme 27, ubiquitin thioesterase 27 and USP27X, is a deubiquitinating enzyme which is mainly characterized for cleaving ubiquitin (Ub) from proteins and other molecules. Ubiquitin binds to proteins so as to regulate the degradation of them via the proteasome and lysosome among many other functions.











