
Yeasts are eukaryotic, single-celled microorganisms classified as members of the fungus kingdom. The first yeast originated hundreds of millions of years ago, and at least 1,500 species are currently recognized. They are estimated to constitute 1% of all described fungal species.

A DNA vaccine is a type of vaccine that transfects a specific antigen-coding DNA sequence into the cells of an organism as a mechanism to induce an immune response.
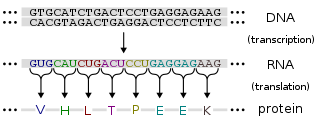
Protein production is the biotechnological process of generating a specific protein. It is typically achieved by the manipulation of gene expression in an organism such that it expresses large amounts of a recombinant gene. This includes the transcription of the recombinant DNA to messenger RNA (mRNA), the translation of mRNA into polypeptide chains, which are ultimately folded into functional proteins and may be targeted to specific subcellular or extracellular locations.
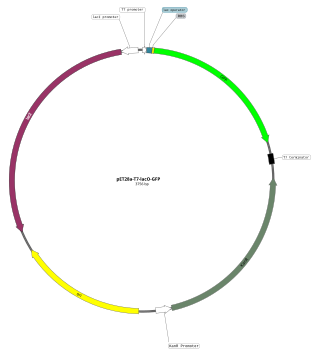
An expression vector, otherwise known as an expression construct, is usually a plasmid or virus designed for gene expression in cells. The vector is used to introduce a specific gene into a target cell, and can commandeer the cell's mechanism for protein synthesis to produce the protein encoded by the gene. Expression vectors are the basic tools in biotechnology for the production of proteins.
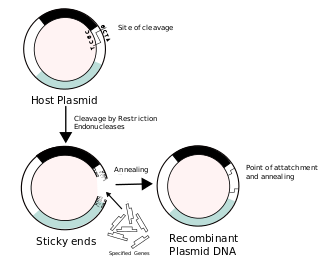
Recombinant DNA (rDNA) molecules are DNA molecules formed by laboratory methods of genetic recombination that bring together genetic material from multiple sources, creating sequences that would not otherwise be found in the genome.
Pharming, a portmanteau of farming and pharmaceutical, refers to the use of genetic engineering to insert genes that code for useful pharmaceuticals into host animals or plants that would otherwise not express those genes, thus creating a genetically modified organism (GMO). Pharming is also known as molecular farming, molecular pharming, or biopharming.

Komagataella is a methylotrophic yeast within the order Saccharomycetales. It was found in the 1960s as Pichia pastoris, with its feature of using methanol as a source of carbon and energy. In 1995, P. pastoris was reassigned into the sole representative of genus Komagataella, becoming Komagataella phaffii. Later studies have further distinguished new species in this genus, resulting in a total of 7 recognized species. It is not uncommon to see the old name still in use in the context of protein production, as of 2023; in less formal use, the yeast may confusingly be referred to as pichia.
Industrial fermentation is the intentional use of fermentation in manufacturing processes. In addition to the mass production of fermented foods and drinks, industrial fermentation has widespread applications in chemical industry. Commodity chemicals, such as acetic acid, citric acid, and ethanol are made by fermentation. Moreover, nearly all commercially produced industrial enzymes, such as lipase, invertase and rennet, are made by fermentation with genetically modified microbes. In some cases, production of biomass itself is the objective, as is the case for single-cell proteins, baker's yeast, and starter cultures for lactic acid bacteria used in cheesemaking.
Microbial genetics is a subject area within microbiology and genetic engineering. Microbial genetics studies microorganisms for different purposes. The microorganisms that are observed are bacteria and archaea. Some fungi and protozoa are also subjects used to study in this field. The studies of microorganisms involve studies of genotype and expression system. Genotypes are the inherited compositions of an organism. Genetic Engineering is a field of work and study within microbial genetics. The usage of recombinant DNA technology is a process of this work. The process involves creating recombinant DNA molecules through manipulating a DNA sequence. That DNA created is then in contact with a host organism. Cloning is also an example of genetic engineering.

Ogataea polymorpha is a methylotrophic yeast with unusual characteristics. It is used as a protein factory for pharmaceuticals.
Arxula adeninivorans is a dimorphic yeast with unusual characteristics. The first description of A. adeninivorans was provided in the mid-eighties. The species was initially designated as Trichosporon adeninovorans. After the first identification in the Netherlands, strains of this species were later on also found in Siberia and in South Africa in soil and in wood hydrolysates. Recently, A. adeninivorans was renamed as Blastobotrys adeninivorans after a detailed phylogenetic comparison with other related yeast species. However, many scientists desire to maintain the popular name A. adeninivorans.
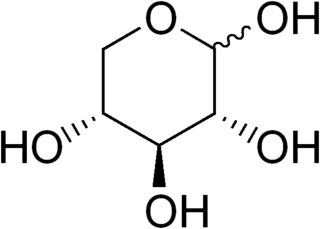
D-Xylose is a five-carbon aldose that can be catabolized or metabolized into useful products by a variety of organisms.
In molecular cloning, a vector is any particle used as a vehicle to artificially carry a foreign nucleic sequence – usually DNA – into another cell, where it can be replicated and/or expressed. A vector containing foreign DNA is termed recombinant DNA. The four major types of vectors are plasmids, viral vectors, cosmids, and artificial chromosomes. Of these, the most commonly used vectors are plasmids. Common to all engineered vectors are an origin of replication, a multicloning site, and a selectable marker.
A killer yeast is a yeast, such as Saccharomyces cerevisiae, which is able to secrete one of a number of toxic proteins which are lethal to susceptible cells. These "killer toxins" are polypeptides that kill sensitive cells of the same or related species, often functioning by creating pores in target cell membranes. These yeast cells are immune to the toxic effects of the protein due to an intrinsic immunity. Killer yeast strains can be a problem in commercial processing because they can kill desirable strains. The killer yeast system was first described in 1963. Study of killer toxins helped to better understand the secretion pathway of yeast, which is similar to those of more complex eukaryotes. It also can be used in treatment of some diseases, mainly those caused by fungi.
A subunit vaccine is a vaccine that contains purified parts of the pathogen that are antigenic, or necessary to elicit a protective immune response. Subunit vaccine can be made from dissembled viral particles in cell culture or recombinant DNA expression, in which case it is a recombinant subunit vaccine.

Genetically modified bacteria were the first organisms to be modified in the laboratory, due to their simple genetics. These organisms are now used for several purposes, and are particularly important in producing large amounts of pure human proteins for use in medicine.
Heterologous expression refers to the expression of a gene or part of a gene in a host organism that does not naturally have the gene or gene fragment in question. Insertion of the gene in the heterologous host is performed by recombinant DNA technology. The purpose of heterologous expression is often to determine the effects of mutations and differential interactions on protein function. It provides an easy path to efficiently express and experiment with combinations of genes and mutants that do not naturally occur.

Molecular cloning is a set of experimental methods in molecular biology that are used to assemble recombinant DNA molecules and to direct their replication within host organisms. The use of the word cloning refers to the fact that the method involves the replication of one molecule to produce a population of cells with identical DNA molecules. Molecular cloning generally uses DNA sequences from two different organisms: the species that is the source of the DNA to be cloned, and the species that will serve as the living host for replication of the recombinant DNA. Molecular cloning methods are central to many contemporary areas of modern biology and medicine.

The role of yeast in winemaking is the most important element that distinguishes wine from fruit juice. In the absence of oxygen, yeast converts the sugars of the fruit into alcohol and carbon dioxide through the process of fermentation. The more sugars in the grapes, the higher the potential alcohol level of the wine if the yeast are allowed to carry out fermentation to dryness. Sometimes winemakers will stop fermentation early in order to leave some residual sugars and sweetness in the wine such as with dessert wines. This can be achieved by dropping fermentation temperatures to the point where the yeast are inactive, sterile filtering the wine to remove the yeast or fortification with brandy or neutral spirits to kill off the yeast cells. If fermentation is unintentionally stopped, such as when the yeasts become exhausted of available nutrients and the wine has not yet reached dryness, this is considered a stuck fermentation.
Transient expression, more frequently referred to "transient gene expression", is the temporary expression of genes that are expressed for a short time after nucleic acid, most frequently plasmid DNA encoding an expression cassette, has been introduced into eukaryotic cells with a chemical delivery agent like calcium phosphate (CaPi) or polyethyleneimine (PEI). However, unlike "stable expression," the foreign DNA does not fuse with the host cell DNA, resulting in the inevitable loss of the vector after several cell replication cycles. The majority of transient gene expressions are done with cultivated animal cells. The technique is also used in plant cells; however, the transfer of nucleic acids into these cells requires different methods than those with animal cells. In both plants and animals, transient expression should result in a time-limited use of transferred nucleic acids, since any long-term expression would be called "stable expression."











