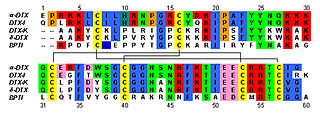
Dendrotoxins are a class of presynaptic neurotoxins produced by mamba snakes (Dendroaspis) that block particular subtypes of voltage-gated potassium channels in neurons, thereby enhancing the release of acetylcholine at neuromuscular junctions. Because of their high potency and selectivity for potassium channels, dendrotoxins have proven to be extremely useful as pharmacological tools for studying the structure and function of these ion channel proteins.

Signal recognition particle (SRP) receptor, also called the docking protein, is a dimer composed of 2 different subunits that are associated exclusively with the rough ER in mammalian cells. Its main function is to identify the SRP units. SRP is a molecule that helps the ribosome-mRNA-polypeptide complexes to settle down on the membrane of the endoplasmic reticulum.

Alpha 1-antichymotrypsin is an alpha globulin glycoprotein that is a member of the serpin superfamily. In humans, it is encoded by the SERPINA3 gene.

ENA/VASP homology proteins or EVH proteins are a family of closely related proteins involved in cell motility in vertebrate and invertebrate animals. EVH proteins are modular proteins that are involved in actin polymerization, as well as interactions with other proteins. Within the cell, Ena/VASP proteins are found at the leading edge of Lamellipodia and at the tips of filopodia. Ena, the founding member of the family was discovered in a drosophila genetic screen for mutations that act as dominant suppressors of the abl non receptor tyrosine kinase. Invertebrate animals have one Ena homologue, whereas mammals have three, named Mena, VASP, and Evl.

Amyloid-like protein 2, also known as APLP2, is a protein that in humans is encoded by the APLP2 gene. APLP2 along with APLP1 are important modulators of glucose and insulin homeostasis.

Latrophilin 1 is a protein that in humans is encoded by the ADGRL1 gene. It is a member of the adhesion-GPCR family of receptors. Family members are characterized by an extended extracellular region with a variable number of protein domains coupled to a TM7 domain via a domain known as the GPCR-Autoproteolysis INducing (GAIN) domain.

Probable G-protein coupled receptor 124 is a protein that in humans is encoded by the GPR124 gene. It is a member of the adhesion-GPCR family of receptors. Family members are characterized by an extended extracellular region with a variable number of protein domains coupled to a TM7 domain via a domain known as the GPCR-Autoproteolysis INducing (GAIN) domain.
Basal cell adhesion molecule, also known as Lutheran antigen, is a plasma membrane glycoprotein that in humans is encoded by the BCAM gene. BCAM has also recently been designated CD239.

Collagen alpha-1(XIII) chain is a protein that in humans is encoded by the COL13A1 gene.

DnaJ homolog subfamily C member 1 is a protein that in humans is encoded by the DNAJC1 gene.
The Kelch motif is a region of protein sequence found widely in proteins from bacteria and eukaryotes. This sequence motif is composed of about 50 amino acid residues which form a structure of a four stranded beta-sheet "blade". This sequence motif is found in between five and eight tandem copies per protein which fold together to form a larger circular solenoid structure called a beta-propeller domain.

GoLoco motif is a protein structural motif.
In molecular biology, the Acyl-CoA-binding protein (ACBP) is a small protein that binds medium- and long-chain acyl-CoA esters with very high affinity and may function as an intracellular carrier of acyl-CoA esters. ACBP is also known as diazepam binding inhibitor (DBI) or endozepine (EP) because of its ability to displace diazepam from the benzodiazepine (BZD) recognition site located on the GABA type A receptor. It is therefore possible that this protein also acts as a neuropeptide to modulate the action of the GABA receptor.
WH1 domain is an evolutionary conserved protein domain. Therefore, it has an important function.

In molecular biology, the AMMECR1 protein is a protein encoded by the AMMECR1 gene on human chromosome Xq22.3.
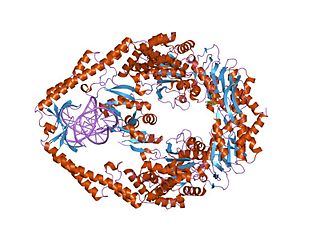
MutS is a mismatch DNA repair protein, originally described in Escherichia coli.

In molecular biology, the BSD domain is an approximately 60-amino-acid-long protein domain named after the BTF2-like transcription factors, synapse-associated proteins and DOS2-like proteins in which it is found. It is also found in several hypothetical proteins. It occurs in one or two copies in a variety of species ranging from primal protozoan to human, and can be found associated with other domains such as the BTB domain or the U-box in multidomain proteins. Its function is as yet unknown.
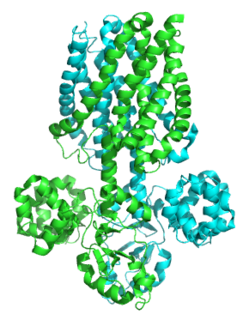
Magnesium transporters E (MgtE) are a family of transmembrane eubacterial MgtE magnesium transporters. Related regions are found also in archaeal and eukaryotic proteins. They have sizes that vary considerably from 311 residues for the Methanococcus thermoautotrophicum protein, 463 residues for a Synechocystis homologue, and 513 residues for the human homologue, SLC41A1. These proteins are capable of transporting Mg2+ and Co2+ but not Ni2+. Multiple alignments contain two highly conserved aspartates that may be involved in cation binding.
YedZ of E. coli has been examined topologically and has 6 transmembrane segments (TMSs) with both the N- and C-termini localized to the cytoplasm. von Rozycki et al. 2004 identified homologues of YedZ in bacteria and animals. YedZ homologues exhibit conserved histidyl residues in their transmembrane domains that may function in heme binding. Some of the homologues encoded in the genomes of various bacteria have YedZ domains fused to transport, electron transfer and biogenesis proteins. One of the animal homologues is the 6 TMS epithelial plasma membrane antigen of the prostate (STAMP1) that is over-expressed in prostate cancer. Some animal homologues have YedZ domains fused C-terminal to homologues of NADP oxidoreductases.
N-glycosyltransferase is an enzyme in prokaryotes which transfers individual hexoses onto asparagine sidechains in substrate proteins, using a nucleotide-bound intermediary, within the cytoplasm. They are distinct from regular N-glycosylating enzymes, which are oligosaccharyltransferases that transfer pre-assembled oligosaccharides. Both enzyme families however target a shared amino acid sequence asparagine—-any amino acid except proline—serine or threonine (N–x–S/T), with some variations.

















