In molecular biology, a riboswitch is a regulatory segment of a messenger RNA molecule that binds a small molecule, resulting in a change in production of the proteins encoded by the mRNA. Thus, an mRNA that contains a riboswitch is directly involved in regulating its own activity, in response to the concentrations of its effector molecule. The discovery that modern organisms use RNA to bind small molecules, and discriminate against closely related analogs, expanded the known natural capabilities of RNA beyond its ability to code for proteins, catalyze reactions, or to bind other RNA or protein macromolecules.
Magnesium transporters are proteins that transport magnesium across the cell membrane. All forms of life require magnesium, yet the molecular mechanisms of Mg2+ uptake from the environment and the distribution of this vital element within the organism are only slowly being elucidated.
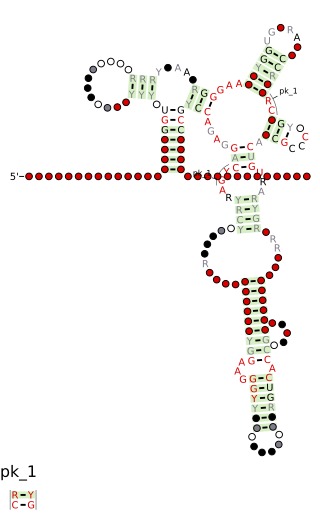
Cobalamin riboswitch is a cis-regulatory element which is widely distributed in 5' untranslated regions of vitamin B12 (Cobalamin) related genes in bacteria.

The FMN riboswitch is a highly conserved RNA element which is naturally occurring, and is found frequently in the 5'-untranslated regions of prokaryotic mRNAs that encode for flavin mononucleotide (FMN) biosynthesis and transport proteins. This element is a metabolite-dependent riboswitch that directly binds FMN in the absence of proteins, thus giving it the ability to regulate RNA expression by responding to changes in the concentration of FMN. In Bacillus subtilis, previous studies have shown that this bacterium utilizes at least two FMN riboswitches, where one controls translation initiation, and the other controls premature transcription termination. Regarding the second riboswitch in Bacilius subtilis, premature transcription termination occurs within the 5' untranslated region of the ribDEAHT operon, precluding access to the ribosome-binding site of ypaA mRNA. FMN riboswitches also have various magnesium and potassium ions dispersed throughout the nucleotide structure, some of which participate in binding of FMN.

The YdaO/YuaA leader is a conserved RNA structure found upstream of the ydaO and yuaA genes in Bacillus subtilis and related genes in other bacteria. Its secondary structure and gene associations were predicted by bioinformatics.

The Lysine riboswitch is a metabolite binding RNA element found within certain messenger RNAs that serve as a precision sensor for the amino acid lysine. Allosteric rearrangement of mRNA structure is mediated by ligand binding, and this results in modulation of gene expression. Lysine riboswitch are most abundant in Bacillota and Gammaproteobacteria where they are found upstream of a number of genes involved in lysine biosynthesis, transport and catabolism. The lysine riboswitch has also been identified independently and called the L box.
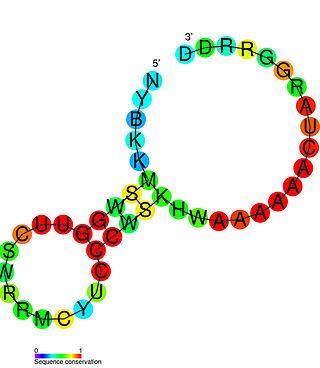
The PreQ1-I riboswitch is a cis-acting element identified in bacteria which regulates expression of genes involved in biosynthesis of the nucleoside queuosine (Q) from GTP. PreQ1 (pre-queuosine1) is an intermediate in the queuosine pathway, and preQ1 riboswitch, as a type of riboswitch, is an RNA element that binds preQ1. The preQ1 riboswitch is distinguished by its unusually small aptamer, compared to other riboswitches. Its atomic-resolution three-dimensional structure has been determined, with the PDB ID 2L1V.

A purine riboswitch is a sequence of ribonucleotides in certain messenger RNA (mRNA) that selectively binds to purine ligands via a natural aptamer domain. This binding causes a conformational change in the mRNA that can affect translation by revealing an expression platform for a downstream gene, or by forming a translation-terminating stem-loop. The ultimate effects of such translational regulation often take action to manage an abundance of the instigating purine, and might produce proteins that facilitate purine metabolism or purine membrane uptake.

The SAM riboswitch is found upstream of a number of genes which code for proteins involved in methionine or cysteine biosynthesis in Gram-positive bacteria. Two SAM riboswitches in Bacillus subtilis that were experimentally studied act at the level of transcription termination control. The predicted secondary structure consists of a complex stem-loop region followed by a single stem-loop terminator region. An alternative and mutually exclusive form involves bases in the 3' segment of helix 1 with those in the 5' region of helix 5 to form a structure termed the anti-terminator form. When SAM is unbound, the anti-terminator sequence sequesters the terminator sequence so the terminator is unable to form, allowing the polymerase to read-through the downstream gene. When S-Adenosyl methionine (SAM) is bound to the aptamer, the anti-terminator is sequestered by an anti-anti-terminator; the terminator forms and terminates the transcription. However, many SAM riboswitches are likely to regulate gene expression at the level of translation.

Usually found in gram-positive bacteria, the T box leader sequence is an RNA element that controls gene expression through the regulation of translation by binding directly to a specific tRNA and sensing its aminoacylation state. This interaction controls expression of downstream aminoacyl-tRNA synthetase genes, amino acid biosynthesis, and uptake-related genes in a negative feedback loop. The uncharged tRNA acts as the effector for transcription antitermination of genes in the T-box leader family. The anticodon of a specific tRNA base pairs to a specifier sequence within the T-box motif, and the NCCA acceptor tail of the tRNA base pairs to a conserved bulge in the T-box antiterminator hairpin.

The ykkC/yxkD leader is a conserved RNA structure found upstream of the ykkC and yxkD genes in Bacillus subtilis and related genes in other bacteria. The function of this family is unclear for many years although it has been suggested that it may function to switch on efflux pumps and detoxification systems in response to harmful environmental molecules. The Thermoanaerobacter tengcongensis sequence AE013027 overlaps with that of purine riboswitch suggesting that the two riboswitches may work in conjunction to regulate the upstream gene which codes for TTE0584 (Q8RC62), a member of the permease family.

This family is a putative regulatory RNA structure that is found upstream of the ylbH gene in B. subtilis and related low GC Gram-positive bacteria.
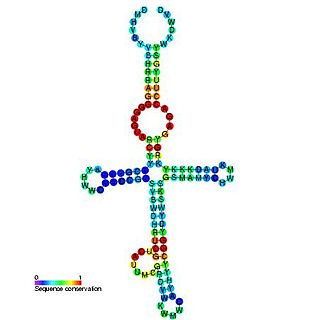
The yybP-ykoY leader RNA element was originally discovered in E. coli during a large scale screen and was named SraF. This family was later found to exist upstream of related families of protein genes in many bacteria, including the yybP and ykoY genes in B. subtilis. The specific functions of these proteins are unknown, but this structured RNA element may be involved in their genetic regulation as a riboswitch. The yybP-ykoY element was later proposed to be manganese-responsive after another associated family of genes, YebN/MntP, was shown to encode Mn2+ efflux pumps in several bacteria. Genetic data and a crystal structure confirmed that yybp-ykoY is a manganese riboswitch that directly binds Mn2+
The Magnesium responsive RNA element, not to be confused with the completely distinct M-box riboswitch, is a cis-regulatory element that regulates the expression of the magnesium transporter protein MgtA. It is located in the 5' UTR of this gene. The mechanism for the potential magnesium-sensing capacity of this RNA is still unclear, though a recent report suggests that the RNA element targets the mgtA transcript for degradation by RNase E when cells are grown in high Mg2+ environments.

The c4 antisense RNA is a non-coding RNA used by certain phages that infect bacteria. It was initially identified in the P1 and P7 phages of E. coli. The identification of c4 antisense RNAs solved the mystery of the mechanism for regulation of the ant gene, which is an anti-repressor.

The fluoride riboswitch is a conserved RNA structure identified by bioinformatics in a wide variety of bacteria and archaea. These RNAs were later shown to function as riboswitches that sense fluoride ions. These "fluoride riboswitches" increase expression of downstream genes when fluoride levels are elevated, and the genes are proposed to help mitigate the toxic effects of very high levels of fluoride.

The yjdF RNA motif is a conserved RNA structure identified using bioinformatics. Most yjdF RNAs are located in bacteria classified within the phylum Bacillota. A yjdF RNA is found in the presumed 5' untranslated region of the yjdF gene in Bacillus subtilis, and almost all yjdF RNAs are found in the 5' UTRs of homologs of this gene. The function of the yjdF gene is unknown, but the protein that it is predicted to encode is classified by the Pfam Database as DUF2992.
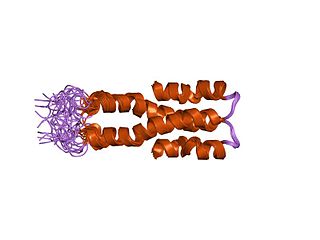
Histone-like nucleoid-structuring protein (H-NS), is one of twelve nucleoid-associated proteins (NAPs) whose main function is the organization of genetic material, including the regulation of gene expression via xenogeneic silencing. H-NS is characterized by an N-terminal domain (NTD) consisting of two dimerization sites, a linker region that is unstructured and a C-terminal domain (CTD) that is responsible for DNA-binding. Though it is a small protein, it provides essential nucleoid compaction and regulation of genes and is highly expressed, functioning as a dimer or multimer. Change in temperature causes H-NS to be dissociated from the DNA duplex, allowing for transcription by RNA polymerase, and in specific regions lead to pathogenic cascades in enterobacteria such as Escherichia coli and the four Shigella species.
The AAC/AAD 5' leader is a disputed genetic element that was proposed to be a conserved RNA structure that is found upstream of the bacterial aminoglycosides antibiotic-resistant genes and that functions as an aminoglycoside-specific riboswitch. The putative RNA is upstream of aminoglycoside acetyl transferase (AAC) and aminoglycoside adenyl transferase (AAD) genes.

The terC RNA motif is a conserved RNA structure that was discovered by bioinformatics. terC motif RNAs are found in Pseudomonadota, within the sub-lineages Alphaproteobacteria and Pseudomonadales.