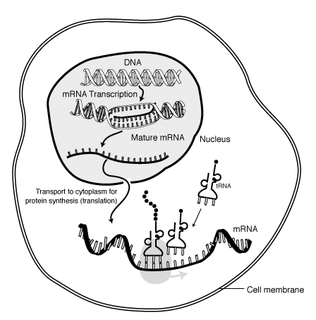
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
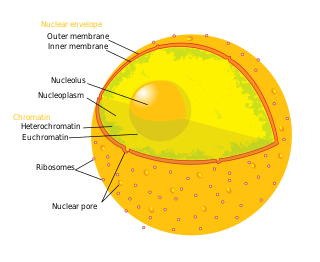
The nucleolus is the largest structure in the nucleus of eukaryotic cells. It is best known as the site of ribosome biogenesis. Nucleoli also participate in the formation of signal recognition particles and play a role in the cell's response to stress. Nucleoli are made of proteins, DNA and RNA and form around specific chromosomal regions called nucleolar organizing regions. Malfunction of nucleoli can be the cause of several human conditions called "nucleolopathies" and the nucleolus is being investigated as a target for cancer chemotherapy.

Ribonucleic acid (RNA) is a polymeric molecule essential in various biological roles in coding, decoding, regulation and expression of genes. RNA and deoxyribonucleic acid (DNA) are nucleic acids. Along with lipids, proteins, and carbohydrates, nucleic acids constitute one of the four major macromolecules essential for all known forms of life. Like DNA, RNA is assembled as a chain of nucleotides, but unlike DNA, RNA is found in nature as a single strand folded onto itself, rather than a paired double strand. Cellular organisms use messenger RNA (mRNA) to convey genetic information that directs synthesis of specific proteins. Many viruses encode their genetic information using an RNA genome.

A reverse transcriptase (RT) is an enzyme used to generate complementary DNA (cDNA) from an RNA template, a process termed reverse transcription. Reverse transcriptases are used by viruses such as HIV and hepatitis B to replicate their genomes, by retrotransposon mobile genetic elements to proliferate within the host genome, and by eukaryotic cells to extend the telomeres at the ends of their linear chromosomes. Contrary to a widely held belief, the process does not violate the flows of genetic information as described by the classical central dogma, as transfers of information from RNA to DNA are explicitly held possible.

A microRNA is a small single-stranded non-coding RNA molecule found in plants, animals and some viruses, that functions in RNA silencing and post-transcriptional regulation of gene expression. miRNAs function via base-pairing with complementary sequences within mRNA molecules. As a result, these mRNA molecules are silenced, by one or more of the following processes: (1) cleavage of the mRNA strand into two pieces, (2) destabilization of the mRNA through shortening of its poly(A) tail, and (3) less efficient translation of the mRNA into proteins by ribosomes.

Transcription is the process of copying a segment of DNA into RNA. The segments of DNA transcribed into RNA molecules that can encode proteins are said to produce messenger RNA (mRNA). Other segments of DNA are copied into RNA molecules called non-coding RNAs (ncRNAs). Averaged over multiple cell types in a given tissue, the quantity of mRNA is more than 10 times the quantity of ncRNA. The general preponderance of mRNA in cells is valid even though less than 2% of the human genome can be transcribed into mRNA, while at least 80% of mammalian genomic DNA can be actively transcribed, with the majority of this 80% considered to be ncRNA.

A non-coding RNA (ncRNA) is an RNA molecule that is not translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally important types of non-coding RNAs include transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), as well as small RNAs such as microRNAs, siRNAs, piRNAs, snoRNAs, snRNAs, exRNAs, scaRNAs and the long ncRNAs such as Xist and HOTAIR.
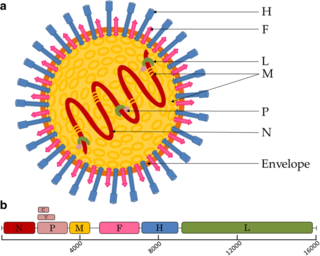
Paramyxoviridae is a family of negative-strand RNA viruses in the order Mononegavirales. Vertebrates serve as natural hosts. Diseases associated with this family include measles, mumps, and respiratory tract infections. The family has four subfamilies, 17 genera, and 78 species, three genera of which are unassigned to a subfamily.

Rhabdoviridae is a family of negative-strand RNA viruses in the order Mononegavirales. Vertebrates, invertebrates, and plants serve as natural hosts. Diseases associated with member viruses include rabies encephalitis caused by the rabies virus, and flu-like symptoms in humans caused by vesiculoviruses. The name is derived from Ancient Greek rhabdos, meaning rod, referring to the shape of the viral particles. The family has 40 genera, most assigned to three subfamilies.

Ribonuclease is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within the EC 2.7 and 3.1 classes of enzymes.
Cauliflower mosaic virus (CaMV) is a member of the genus Caulimovirus, one of the six genera in the family Caulimoviridae, which are pararetroviruses that infect plants. Pararetroviruses replicate through reverse transcription just like retroviruses, but the viral particles contain DNA instead of RNA.
The 5′ untranslated region is the region of a messenger RNA (mRNA) that is directly upstream from the initiation codon. This region is important for the regulation of translation of a transcript by differing mechanisms in viruses, prokaryotes and eukaryotes. While called untranslated, the 5′ UTR or a portion of it is sometimes translated into a protein product. This product can then regulate the translation of the main coding sequence of the mRNA. In many organisms, however, the 5′ UTR is completely untranslated, instead forming complex secondary structure to regulate translation.

CRISPR is a family of DNA sequences found in the genomes of prokaryotic organisms such as bacteria and archaea. These sequences are derived from DNA fragments of bacteriophages that had previously infected the prokaryote. They are used to detect and destroy DNA from similar bacteriophages during subsequent infections. Hence these sequences play a key role in the antiviral defense system of prokaryotes and provide a form of acquired immunity. CRISPR are found in approximately 50% of sequenced bacterial genomes and nearly 90% of sequenced archaea.
Post-transcriptional modification or co-transcriptional modification is a set of biological processes common to most eukaryotic cells by which an RNA primary transcript is chemically altered following transcription from a gene to produce a mature, functional RNA molecule that can then leave the nucleus and perform any of a variety of different functions in the cell. There are many types of post-transcriptional modifications achieved through a diverse class of molecular mechanisms.
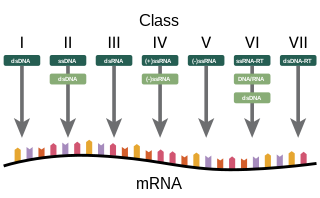
Baltimore classification is a system used to classify viruses based on their manner of messenger RNA (mRNA) synthesis. By organizing viruses based on their manner of mRNA production, it is possible to study viruses that behave similarly as a distinct group. Seven Baltimore groups are described that take into consideration whether the viral genome is made of deoxyribonucleic acid (DNA) or ribonucleic acid (RNA), whether the genome is single- or double-stranded, and whether the sense of a single-stranded RNA genome is positive or negative.
Small nucleolar RNAs (snoRNAs) are a class of small RNA molecules that primarily guide chemical modifications of other RNAs, mainly ribosomal RNAs, transfer RNAs and small nuclear RNAs. There are two main classes of snoRNA, the C/D box snoRNAs, which are associated with methylation, and the H/ACA box snoRNAs, which are associated with pseudouridylation. SnoRNAs are commonly referred to as guide RNAs but should not be confused with the guide RNAs that direct RNA editing in trypanosomes.
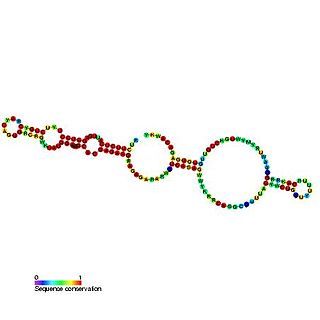
U23 belongs to the H/ACA class of snoRNAs. snoRNAs bind a number of proteins to form snoRNP complexes. This class are thought to guide the sites of modification of uridines to pseudouridines by forming direct base pairing interactions with substrate RNAs. Targets include ribosomal and spliceosomal RNAs as well as the Trypanosoma spliced leader RNA as possibly other, still unknown cellular RNAs. U23 can direct the pseudouridylation of U97 in human 18S rRNA. U23 is encoded within intron 12 of the nucleolin gene in human, mouse, rat chicken, and Xenopus laevis.
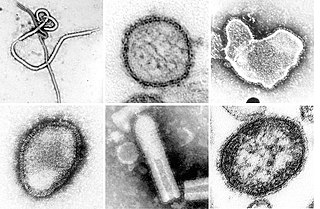
Negative-strand RNA viruses are a group of related viruses that have negative-sense, single-stranded genomes made of ribonucleic acid. They have genomes that act as complementary strands from which messenger RNA (mRNA) is synthesized by the viral enzyme RNA-dependent RNA polymerase (RdRp). During replication of the viral genome, RdRp synthesizes a positive-sense antigenome that it uses as a template to create genomic negative-sense RNA. Negative-strand RNA viruses also share a number of other characteristics: most contain a viral envelope that surrounds the capsid, which encases the viral genome, -ssRNA virus genomes are usually linear, and it is common for their genome to be segmented.

A ribonucleic acid (RNA) vaccine or messenger RNA (mRNA) vaccine is a type of vaccine that uses a copy of a molecule called messenger RNA (mRNA) to produce an immune response. The vaccine transfects molecules of synthetic RNA into immune cells, where the vaccine functions as mRNA, causing the cells to build foreign protein that would normally be produced by a pathogen or by a cancer cell. These protein molecules stimulate an adaptive immune response that teaches the body to identify and destroy the corresponding pathogen or cancer cells. The mRNA is delivered by a co-formulation of the RNA encapsulated in lipid nanoparticles that protect the RNA strands and help their absorption into the cells.

Katalin Karikó is a Hungarian biochemist who specializes in RNA-mediated mechanisms. Her research has been the development of in vitro-transcribed mRNA for protein therapies. She co-founded and was CEO of RNARx, from 2006 to 2013. Since 2013, she has been associated with BioNTech RNA Pharmaceuticals, first as a vice president and promoted to senior vice president in 2019. She also is an adjunct professor at the University of Pennsylvania.
















