In molecular biology, biosynthesis is a multi-step, enzyme-catalyzed process where substrates are converted into more complex products in living organisms. In biosynthesis, simple compounds are modified, converted into other compounds, or joined to form macromolecules. This process often consists of metabolic pathways. Some of these biosynthetic pathways are located within a single cellular organelle, while others involve enzymes that are located within multiple cellular organelles. Examples of these biosynthetic pathways include the production of lipid membrane components and nucleotides. Biosynthesis is usually synonymous with anabolism.

An antimetabolite is a chemical that inhibits the use of a metabolite, which is another chemical that is part of normal metabolism. Such substances are often similar in structure to the metabolite that they interfere with, such as the antifolates that interfere with the use of folic acid; thus, competitive inhibition can occur, and the presence of antimetabolites can have toxic effects on cells, such as halting cell growth and cell division, so these compounds are used in chemotherapy for cancer.
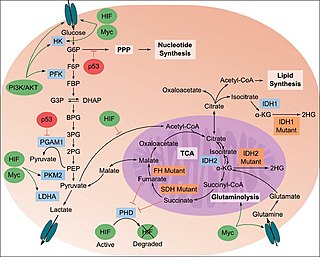
The study of the tumor metabolism, also known as tumor metabolome describes the different characteristic metabolic changes in tumor cells. The characteristic attributes of the tumor metabolome are high glycolytic enzyme activities, the expression of the pyruvate kinase isoenzyme type M2, increased channeling of glucose carbons into synthetic processes, such as nucleic acid, amino acid and phospholipid synthesis, a high rate of pyrimidine and purine de novo synthesis, a low ratio of Adenosine triphosphate and Guanosine triphosphate to Cytidine triphosphate and Uridine triphosphate, low Adenosine monophosphate levels, high glutaminolytic capacities, release of immunosuppressive substances and dependency on methionine.

Amino acid biosynthesis is the set of biochemical processes by which the amino acids are produced. The substrates for these processes are various compounds in the organism's diet or growth media. Not all organisms are able to synthesize all amino acids. For example, humans can synthesize 11 of the 20 standard amino acids. These 11 are called the non-essential amino acids).
Pyrimidine biosynthesis occurs both in the body and through organic synthesis.

CAD protein is a trifunctional multi-domain enzyme involved in the first three steps of pyrimidine biosynthesis. De-novo synthesis starts with cytosolic carbamoylphosphate synthetase II which uses glutamine, carbon dioxide and ATP. This enzyme is inhibited by uridine triphosphate.
CTP synthase is an enzyme involved in pyrimidine biosynthesis that interconverts UTP and CTP.
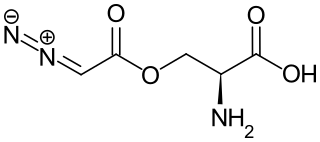
Azaserine is a naturally occurring serine derivative diazo compound with antineoplastic and antibiotic properties deriving from its action as a purinergic antagonist and structural similarity to glutamine. Azaserine acts by competitively inhibiting glutamine amidotransferase, a key enzyme responsible for glutamine metabolism.

Glutaminase is an amidohydrolase enzyme that generates glutamate from glutamine. Glutaminase has tissue-specific isoenzymes. Glutaminase has an important role in glial cells.
Barbiturase is a zinc-containing amidohydrolase. Its systemic name is barbiturate amidohydrolase (3-oxo-3-ureidopropanoate-forming). Barbiturase acts as a catalyst in the second step of oxidative pyrimidine degradation, promoting the ring-opening hydrolysis of barbituric acid to ureidomalonic acid. Although grouped into the naturally existing amidohydrolases, it demonstrates more homology with cyanuric acid amidohydrolase. Therefore, it has been proposed that barbiturase, along with cyanuric acid, should be grouped into a new family. KEGG

Carbamoyl phosphate synthetase catalyzes the ATP-dependent synthesis of carbamoyl phosphate from glutamine or ammonia and bicarbonate. This enzyme catalyzes the reaction of ATP and bicarbonate to produce carboxy phosphate and ADP. Carboxy phosphate reacts with ammonia to give carbamic acid. In turn, carbamic acid reacts with a second ATP to give carbamoyl phosphate plus ADP.
Purine metabolism refers to the metabolic pathways to synthesize and break down purines that are present in many organisms.

Amidophosphoribosyltransferase (ATase), also known as glutamine phosphoribosylpyrophosphate amidotransferase (GPAT), is an enzyme responsible for catalyzing the conversion of 5-phosphoribosyl-1-pyrophosphate (PRPP) into 5-phosphoribosyl-1-amine (PRA), using the amine group from a glutamine side-chain. This is the committing step in de novo purine synthesis. In humans it is encoded by the PPAT gene. ATase is a member of the purine/pyrimidine phosphoribosyltransferase family.

Asparagine synthetase is a chiefly cytoplasmic enzyme that generates asparagine from aspartate. This amidation reaction is similar to that promoted by glutamine synthetase. The enzyme is ubiquitous in its distribution in mammalian organs, but basal expression is relatively low in tissues other than the exocrine pancreas.

In enzymology, an omega-amidase (EC 3.5.1.3) is an enzyme that catalyzes the chemical reaction
In enzymology, a glutamine-pyruvate transaminase is an enzyme that catalyzes the chemical reaction

Uridine-cytidine kinase 2 (UCK2) is an enzyme that in humans is encoded by the UCK2 gene.
Glutaminolysis (glutamine + -lysis) is a series of biochemical reactions by which the amino acid glutamine is lysed to glutamate, aspartate, CO2, pyruvate, lactate, alanine and citrate.
Asparagine synthase (glutamine-hydrolysing) (EC 6.3.5.4, asparagine synthetase (glutamine-hydrolysing), glutamine-dependent asparagine synthetase, asparagine synthetase B, AS, AS-B) is an enzyme with systematic name L-aspartate:L-glutamine amido-ligase (AMP-forming). This enzyme catalyses the following chemical reaction
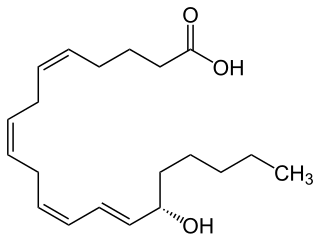
15-Hydroxyeicosatetraenoic acid (also termed 15-HETE, 15(S)-HETE, and 15S-HETE) is an eicosanoid, i.e. a metabolite of arachidonic acid. Various cell types metabolize arachidonic acid to 15(S)-hydroperoxyeicosatetraenoic acid (15(S)-HpETE). This initial hydroperoxide product is extremely short-lived in cells: if not otherwise metabolized, it is rapidly reduced to 15(S)-HETE. Both of these metabolites, depending on the cell type which forms them, can be further metabolized to 15-oxo-eicosatetraenoic acid (15-oxo-ETE), 5(S),15(S)-dihydroxy-eicosatetraenoic acid (5(S),15(S)-diHETE), 5-oxo-15(S)-hydroxyeicosatetraenoic acid (5-oxo-15(S)-HETE), a subset of specialized pro-resolving mediators viz., the lipoxins, a class of pro-inflammatory mediators, the eoxins, and other products that have less well-defined activities and functions. Thus, 15(S)-HETE and 15(S)-HpETE, in addition to having intrinsic biological activities, are key precursors to numerous biologically active derivatives.














