
Ribosomes are macromolecular machines, found within all living cells, that perform biological protein synthesis. Ribosomes link amino acids together in the order specified by the codons of messenger RNA (mRNA) molecules to form polypeptide chains. Ribosomes consist of two major components: the small and large ribosomal subunits. Each subunit consists of one or more ribosomal RNA (rRNA) molecules and many ribosomal proteins. The ribosomes and associated molecules are also known as the translational apparatus.

Ribonuclease H is a family of non-sequence-specific endonuclease enzymes that catalyze the cleavage of RNA in an RNA/DNA substrate via a hydrolytic mechanism. Members of the RNase H family can be found in nearly all organisms, from bacteria to archaea to eukaryotes.

The ATP-binding cassette transporters are a transport system superfamily that is one of the largest and possibly one of the oldest gene families. It is represented in all extant phyla, from prokaryotes to humans.

Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from ribosomal DNA (rDNA) and then bound to ribosomal proteins to form small and large ribosome subunits. rRNA is the physical and mechanical factor of the ribosome that forces transfer RNA (tRNA) and messenger RNA (mRNA) to process and translate the latter into proteins. Ribosomal RNA is the predominant form of RNA found in most cells; it makes up about 80% of cellular RNA despite never being translated into proteins itself. Ribosomes are composed of approximately 60% rRNA and 40% ribosomal proteins by mass.
Bacterial translation is the process by which messenger RNA is translated into proteins in bacteria.
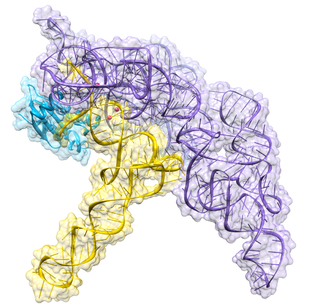
Ribonuclease P is a type of ribonuclease which cleaves RNA. RNase P is unique from other RNases in that it is a ribozyme – a ribonucleic acid that acts as a catalyst in the same way that a protein-based enzyme would. Its function is to cleave off an extra, or precursor, sequence of RNA on tRNA molecules. Further, RNase P is one of two known multiple turnover ribozymes in nature, the discovery of which earned Sidney Altman and Thomas Cech the Nobel Prize in Chemistry in 1989: in the 1970s, Altman discovered the existence of precursor tRNA with flanking sequences and was the first to characterize RNase P and its activity in processing of the 5' leader sequence of precursor tRNA. Recent findings also reveal that RNase P has a new function. It has been shown that human nuclear RNase P is required for the normal and efficient transcription of various small noncoding RNAs, such as tRNA, 5S rRNA, SRP RNA and U6 snRNA genes, which are transcribed by RNA polymerase III, one of three major nuclear RNA polymerases in human cells.
Initiation factors are proteins that bind to the small subunit of the ribosome during the initiation of translation, a part of protein biosynthesis.

Ribosome biogenesis is the process of making ribosomes. In prokaryotes, this process takes place in the cytoplasm with the transcription of many ribosome gene operons. In eukaryotes, it takes place both in the cytoplasm and in the nucleolus. It involves the coordinated function of over 200 proteins in the synthesis and processing of the three prokaryotic or four eukaryotic rRNAs, as well as assembly of those rRNAs with the ribosomal proteins. Most of the ribosomal proteins fall into various energy-consuming enzyme families including ATP-dependent RNA helicases, AAA-ATPases, GTPases, and kinases. About 60% of a cell's energy is spent on ribosome production and maintenance.
Iron-binding proteins are carrier proteins and metalloproteins that are important in iron metabolism and the immune response. Iron is required for life.
A ribosomal protein is any of the proteins that, in conjunction with rRNA, make up the ribosomal subunits involved in the cellular process of translation. A large part of the knowledge about these organic molecules has come from the study of E. coli ribosomes. All ribosomal proteins have been isolated and many specific antibodies have been produced. These, together with electronic microscopy and the use of certain reactives, have allowed for the determination of the topography of the proteins in the ribosome. E. coli, other bacteria and Archaea have a 30S small subunit and a 50S large subunit, whereas humans and yeasts have a 40S small subunit and a 60S large subunit. Equivalent subunits are frequently numbered differently between bacteria, Archaea, yeasts and humans. More recently, a near-complete (near)atomic picture of the ribosomal proteins is emerging from the latest high-resolution cryo-EM data.
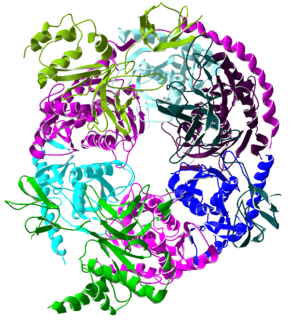
The exosome complex is a multi-protein intracellular complex capable of degrading various types of RNA molecules. Exosome complexes are found in both eukaryotic cells and archaea, while in bacteria a simpler complex called the degradosome carries out similar functions.
Translocase is a general term for a protein that assists in moving another molecule, usually across a cell membrane. These enzymes catalyze the movement of ions or molecules across membranes or their separation within membranes. The reaction is designated as a transfer from “side 1” to “side 2” because the designations “in” and “out”, which had previously been used, can be ambiguous. Translocases are the most common secretion system in Gram positive bacteria.
Leader peptidase A (LepA) is an elongation factor that is thought to back-translocate on the ribosome during the translation of RNA to proteins in all prokaryotes and eukaryotes that have maintained functioning mitochondria. There are three primary elongation factors that are known to be the main contributors to facilitate elongation during protein synthesis; due to LepA's now acknowledged proofreading function in translation, scientists are now lobbying to have LepA renamed as EF-4.
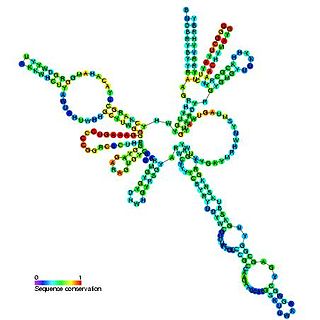
RNase MRP is an enzymatically active ribonucleoprotein with two distinct roles in eukaryotes. RNAse MRP stands for RNAse for mitochondrial RNA processing. In mitochondria it plays a direct role in the initiation of mitochondrial DNA replication. In the nucleus it is involved in precursor rRNA processing, where it cleaves the internal transcribed spacer 1 between 18S and 5.8S rRNAs. Despite distinct functions, RNase MRP has been shown to be evolutionarily related to RNase P. Like eukaryotic RNase P, RNase MRP is not catalytically active without associated protein subunits.
A ribosome binding site, or ribosomal binding site (RBS), is a sequence of nucleotides upstream of the start codon of an mRNA transcript that is responsible for the recruitment of a ribosome during the initiation of translation. Mostly, RBS refers to bacterial sequences, although internal ribosome entry sites (IRES) have been described in mRNAs of eukaryotic cells or viruses that infect eukaryotes. Ribosome recruitment in eukaryotes is generally mediated by the 5' cap present on eukaryotic mRNAs.
The degradosome is a multiprotein complex present in most bacteria that is involved in the processing of ribosomal RNA and the degradation of messenger RNA and is regulated by Non-coding RNA. It contains the proteins RNA helicase B, RNase E and Polynucleotide phosphorylase.

In molecular biology, ATP-binding domain of ABC transporters is a water-soluble domain of transmembrane ABC transporters.

A protein synthesis inhibitor is a compound that stops or slows the growth or proliferation of cells by disrupting the processes that lead directly to the generation of new proteins.
Ribonuclease E is a bacterial ribonuclease that participates in the processing of ribosomal RNA and the chemical degradation of bulk cellular RNA.
Archaeal initiation factors are proteins that are used during the translation step of protein synthesis in archaea. The principal functions these proteins perform include ribosome RNA/mRNA recognition, delivery of the initiator Met-tRNAiMet, methionine bound tRNAi, to the 40s ribosome, and proofreading of the initiation complex.













