
In genetics, complementary DNA (cDNA) is DNA synthesized from a single-stranded RNA template in a reaction catalyzed by the enzyme reverse transcriptase. cDNA is often used to express a specific protein in a cell that does not normally express that protein, or to sequence or quantify mRNA molecules using DNA based methods. cDNA that codes for a specific protein can be transferred to a recipient cell for expression, often bacterial or yeast expression systems. cDNA is also generated to analyze transcriptomic profiles in bulk tissue, single cells, or single nuclei in assays such as microarrays, qPCR, and RNA-seq.

A retrovirus is a type of virus that inserts a DNA copy of its RNA genome into the DNA of a host cell that it invades, thus changing the genome of that cell. After invading a host cell's cytoplasm, the virus uses its own reverse transcriptase enzyme to produce DNA from its RNA genome, the reverse of the usual pattern, thus retro (backwards). The new DNA is then incorporated into the host cell genome by an integrase enzyme, at which point the retroviral DNA is referred to as a provirus. The host cell then treats the viral DNA as part of its own genome, transcribing and translating the viral genes along with the cell's own genes, producing the proteins required to assemble new copies of the virus. Many retroviruses cause serious diseases in humans, other mammals, and birds.

Hepadnaviridae is a family of viruses. Humans, apes, and birds serve as natural hosts. There are currently 18 species in this family, divided among 5 genera. Its best-known member is hepatitis B virus. Diseases associated with this family include: liver infections, such as hepatitis, hepatocellular carcinomas, and cirrhosis. It is the sole accepted family in the order Blubervirales.
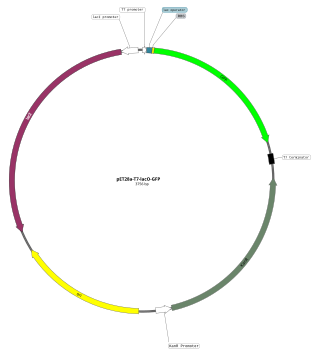
An expression vector, otherwise known as an expression construct, is usually a plasmid or virus designed for gene expression in cells. The vector is used to introduce a specific gene into a target cell, and can commandeer the cell's mechanism for protein synthesis to produce the protein encoded by the gene. Expression vectors are the basic tools in biotechnology for the production of proteins.

Plant viruses are viruses that affect plants. Like all other viruses, plant viruses are obligate intracellular parasites that do not have the molecular machinery to replicate without a host. Plant viruses can be pathogenic to vascular plants.

Geminiviridae is a family of plant viruses that encode their genetic information on a circular genome of single-stranded (ss) DNA. There are 520 species in this family, assigned to 14 genera. Diseases associated with this family include: bright yellow mosaic, yellow mosaic, yellow mottle, leaf curling, stunting, streaks, reduced yields. They have single-stranded circular DNA genomes encoding genes that diverge in both directions from a virion strand origin of replication. According to the Baltimore classification they are considered class II viruses. It is the largest known family of single stranded DNA viruses.
Caulimoviridae is a family of viruses infecting plants. There are 94 species in this family, assigned to 11 genera. Viruses belonging to the family Caulimoviridae are termed double-stranded DNA (dsDNA) reverse-transcribing viruses i.e. viruses that contain a reverse transcription stage in their replication cycle. This family contains all plant viruses with a dsDNA genome that have a reverse transcribing phase in their lifecycle.

Tombusviridae is a family of single-stranded positive sense RNA plant viruses. There are three subfamilies, 17 genera, and 95 species in this family. The name is derived from Tomato bushy stunt virus (TBSV).
Lentivirus is a genus of retroviruses that cause chronic and deadly diseases characterized by long incubation periods, in humans and other mammalian species. The genus includes the human immunodeficiency virus (HIV), which causes AIDS. Lentiviruses are distributed worldwide, and are known to be hosted in apes, cows, goats, horses, cats, and sheep as well as several other mammals.
The genome and proteins of HIV (human immunodeficiency virus) have been the subject of extensive research since the discovery of the virus in 1983. "In the search for the causative agent, it was initially believed that the virus was a form of the Human T-cell leukemia virus (HTLV), which was known at the time to affect the human immune system and cause certain leukemias. However, researchers at the Pasteur Institute in Paris isolated a previously unknown and genetically distinct retrovirus in patients with AIDS which was later named HIV." Each virion comprises a viral envelope and associated matrix enclosing a capsid, which itself encloses two copies of the single-stranded RNA genome and several enzymes. The discovery of the virus itself occurred two years following the report of the first major cases of AIDS-associated illnesses.
Ribosome shunting is a mechanism of translation initiation in which ribosomes bypass, or "shunt over", parts of the 5' untranslated region to reach the start codon. However, a benefit of ribosomal shunting is that it can translate backwards allowing more information to be stored than usual in an mRNA molecule. Some viral RNAs have been shown to use ribosome shunting as a more efficient form of translation during certain stages of viral life cycle or when translation initiation factors are scarce. Some viruses known to use this mechanism include adenovirus, Sendai virus, human papillomavirus, duck hepatitis B pararetrovirus, rice tungro bacilliform viruses, and cauliflower mosaic virus. In these viruses the ribosome is directly translocated from the upstream initiation complex to the start codon (AUG) without the need to unwind RNA secondary structures.
Baltimore classification is a system used to classify viruses based on their manner of messenger RNA (mRNA) synthesis. By organizing viruses based on their manner of mRNA production, it is possible to study viruses that behave similarly as a distinct group. Seven Baltimore groups are described that take into consideration whether the viral genome is made of deoxyribonucleic acid (DNA) or ribonucleic acid (RNA), whether the genome is single- or double-stranded, and whether the sense of a single-stranded RNA genome is positive or negative.

Potyvirus is a genus of positive-strand RNA viruses in the family Potyviridae. Plants serve as natural hosts. Like begomoviruses, members of this genus may cause significant losses in agricultural, pastoral, horticultural, and ornamental crops. More than 200 species of aphids spread potyviruses, and most are from the subfamily Aphidinae. The genus contains 190 species and potyviruses account for about thirty percent of all currently known plant viruses.

A viroplasm, sometimes called "virus factory" or "virus inclusion", is an inclusion body in a cell where viral replication and assembly occurs. They may be thought of as viral factories in the cell. There are many viroplasms in one infected cell, where they appear dense to electron microscopy. Very little is understood about the mechanism of viroplasm formation.
In molecular biology and genetics, the sense of a nucleic acid molecule, particularly of a strand of DNA or RNA, refers to the nature of the roles of the strand and its complement in specifying a sequence of amino acids. Depending on the context, sense may have slightly different meanings. For example, negative-sense strand of DNA is equivalent to the template strand, whereas the positive-sense strand is the non-template strand whose nucleotide sequence is equivalent to the sequence of the mRNA transcript.

Molluscum contagiosum virus (MCV) is a species of DNA poxvirus that causes the human skin infection molluscum contagiosum. Molluscum contagiosum affects about 200,000 people a year, about 1% of all diagnosed skin diseases. Diagnosis is based on the size and shape of the skin lesions and can be confirmed with a biopsy, as the virus cannot be routinely cultured. Molluscum contagiosum virus is the only species in the genus Molluscipoxvirus. MCV is a member of the subfamily Chordopoxvirinae of family Poxviridae. Other commonly known viruses that reside in the subfamily Chordopoxvirinae are variola virus and monkeypox virus.
Group-specific antigen, or gag, is the polyprotein that contains the core structural proteins of an Ortervirus. It was named as such because scientists used to believe it was antigenic. Now it is known that it makes up the inner shell, not the envelope exposed outside. It makes up all the structural units of viral conformation and provides supportive framework for mature virion.

Alfalfa mosaic virus (AMV), also known as Lucerne mosaic virus or Potato calico virus, is a worldwide distributed phytopathogen that can lead to necrosis and yellow mosaics on a large variety of plant species, including commercially important crops. It is the only Alfamovirus of the family Bromoviridae. In 1931 Weimer J.L. was the first to report AMV in alfalfa. Transmission of the virus occurs mainly by some aphids, by seeds or by pollen to the seed.

Riboviria is a realm of viruses that includes all viruses that use a homologous RNA-dependent polymerase for replication. It includes RNA viruses that encode an RNA-dependent RNA polymerase, as well as reverse-transcribing viruses that encode an RNA-dependent DNA polymerase. RNA-dependent RNA polymerase (RdRp), also called RNA replicase, produces RNA from RNA. RNA-dependent DNA polymerase (RdDp), also called reverse transcriptase (RT), produces DNA from RNA. These enzymes are essential for replicating the viral genome and transcribing viral genes into messenger RNA (mRNA) for translation of viral proteins.

Orthornavirae is a kingdom of viruses that have genomes made of ribonucleic acid (RNA), including genes which encode an RNA-dependent RNA polymerase (RdRp). The RdRp is used to transcribe the viral RNA genome into messenger RNA (mRNA) and to replicate the genome. Viruses in this kingdom share a number of characteristics which promote rapid evolution, including high rates of genetic mutation, recombination, and reassortment.
















