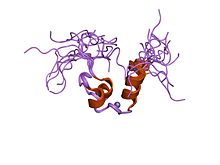
A zinc finger is a small protein structural motif that is characterized by the coordination of one or more zinc ions (Zn2+) in order to stabilize the fold. It was originally coined to describe the finger-like appearance of a hypothesized structure from the African clawed frog (Xenopus laevis) transcription factor IIIA. However, it has been found to encompass a wide variety of differing protein structures in eukaryotic cells. Xenopus laevis TFIIIA was originally demonstrated to contain zinc and require the metal for function in 1983, the first such reported zinc requirement for a gene regulatory protein followed soon thereafter by the Krüppel factor in Drosophila. It often appears as a metal-binding domain in multi-domain proteins.

In molecular biology, heat shock factors (HSF), are the transcription factors that regulate the expression of the heat shock proteins. A typical example is the heat shock factor of Drosophila melanogaster.
POU is a family of proteins that have well-conserved homeodomains.

Doublesex and mab-3 related transcription factor 1, also known as DMRT1, is a protein which in humans is encoded by the DMRT1 gene.

Mediator of RNA polymerase II transcription subunit 26 is an enzyme that in humans is encoded by the MED26 gene. It forms part of the Mediator complex.
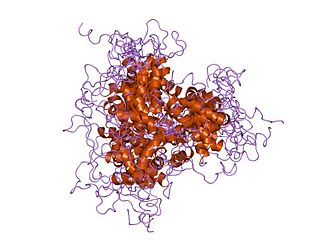
The fork head domain is a type of protein domain that is often found in transcription factors and whose purpose is to bind DNA.

Notch proteins are a family of type-1 transmembrane proteins that form a core component of the Notch signaling pathway, which is highly conserved in metazoans. The Notch extracellular domain mediates interactions with DSL family ligands, allowing it to participate in juxtacrine signaling. The Notch intracellular domain acts as a transcriptional activator when in complex with CSL family transcription factors. Members of this Type 1 transmembrane protein family share several core structures, including an extracellular domain consisting of multiple epidermal growth factor (EGF)-like repeats and an intracellular domain transcriptional activation domain (TAD). Notch family members operate in a variety of different tissues and play a role in a variety of developmental processes by controlling cell fate decisions. Much of what is known about Notch function comes from studies done in Caenorhabditis elegans (C.elegans) and Drosophila melanogaster. Human homologs have also been identified, but details of Notch function and interactions with its ligands are not well known in this context.

Doublesex (dsx) is a gene that is involved in the sex determination system of many insects including the fruit fly Drosophila melanogaster.
The RNA-binding Proteins Database (RBPDB) is a biological database of RNA-binding protein specificities that includes experimental observations of RNA-binding sites. The experimental results included are both in vitro and in vivo from primary literature. It includes four metazoan species, which are Homo sapiens, Mus musculus, Drosophila melanogaster, and Caenorhabditis elegans. RNA-binding domains included in this database are RNA recognition motif, K homology, CCCH zinc finger, and more domains. As of 2021, the latest RBPDB release includes 1,171 RNA-binding proteins.
Mediator of RNA polymerase II transcription subunit 29 (Med29) is a transcription suppressor that in humans is encoded by the MED29 gene. It represents subunit MED29 of the Mediator complex.
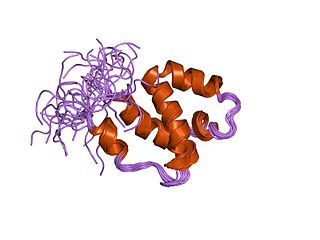
In molecular biology, the protein domain SWAP is derived from the term Suppressor-of-White-APricot, a splicing regulator from the model organism Drosophila melanogaster. The protein domain is found in regulators that control splicing. It is found in splicing regulatory proteins. When a gene is expressed the DNA must be transcribed into messenger RNA (mRNA). However, it sometimes contains intervening or interrupting sequences named introns. mRNA splicing helps to remove these sequences, leaving a more favourable sequence. mRNA splicing is an essential event in the post-transcriptional modification process of gene expression. SWAP helps to control this process in all cells except gametes.

In molecular biology the MYND-type zinc finger domain is a conserved protein domain. The MYND domain is present in a large group of proteins that includes RP-8 (PDCD2), Nervy, and predicted proteins from Drosophila, mammals, Caenorhabditis elegans, yeast, and plants. The MYND domain consists of a cluster of cysteine and histidine residues, arranged with an invariant spacing to form a potential zinc-binding motif. Mutating conserved cysteine residues in the DEAF-1 MYND domain does not abolish DNA binding, which suggests that the MYND domain might be involved in protein-protein interactions. Indeed, the MYND domain of ETO/MTG8 interacts directly with the N-CoR and SMRT co-repressors. Aberrant recruitment of co-repressor complexes and inappropriate transcriptional repression is believed to be a general mechanism of leukemogenesis caused by the t(8;21) translocations that fuse ETO with the acute myelogenous leukemia 1 (AML1) protein. ETO has been shown to be a co-repressor recruited by the promyelocytic leukemia zinc finger (PLZF) protein. A divergent MYND domain present in the adenovirus E1A binding protein BS69 was also shown to interact with N-CoR and mediate transcriptional repression. The current evidence suggests that the MYND motif in mammalian proteins constitutes a protein-protein interaction domain that functions as a co-repressor-recruiting interface.
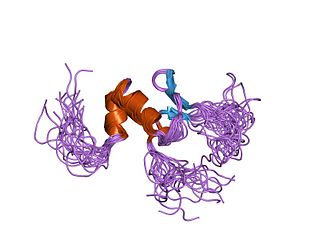
In molecular biology the BED-type zinc finger domain is a protein domain which was named after the Drosophila proteins BEAF and DREF, is found in one or more copies in cellular regulatory factors and transposases from plants, animals and fungi. The BED finger is an about 50 to 60 amino acid residues domain that contains a characteristic motif with two highly conserved aromatic positions, as well as a shared pattern of cysteines and histidines that is predicted to form a zinc finger. As diverse BED fingers are able to bind DNA, it has been suggested that DNA-binding is the general function of this domain. Some proteins known to contain a BED domain include animal, plant and fungi AC1 and Hobo-like transposases; Caenorhabditis elegans Dpy-20 protein, a predicted cuticular gene transcriptional regulator; Drosophila BEAF, thought to be involved in chromatin insulation; Drosophila DREF, a transcriptional regulator for S-phase genes; and tobacco 3AF1 and tomato E4/E8-BP1, light- and ethylene-regulated DNA binding proteins that contain two BED fingers. Most genes in this family are believed to have evolved from the hAT family of DNA transposons.
In molecular biology, the BEN domain is a protein domain which is found in diverse proteins including:
In molecular biology, the BESS domain is a protein domain which has been named after the three proteins that originally defined the domain: BEAF, Suvar(3)7 and Stonewall ). The BESS domain is 40 amino acid residues long and is predicted to be composed of three alpha helices, as such it might be related to the myb/SANT HTH domain. The BESS domain directs a variety of protein-protein interactions, including interactions with itself, with Dorsal, and with a TBP-associated factor. It is found in a single copy in Drosophila proteins and is often associated with the MADF domain.
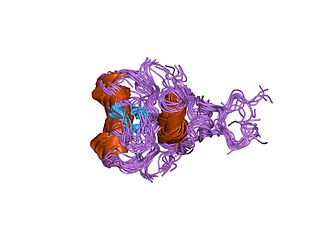
In molecular biology, GATA zinc fingers are zinc-containing domains found in a number of transcription factors. Some members of this class of zinc fingers specifically bind the DNA sequence (A/T)GATA(A/G) in the regulatory regions of genes., giving rise to the name of the domain. In these domains, a single zinc ion is coordinated by 4 cysteine residues. NMR studies have shown the core of the Znf to comprise 2 irregular anti-parallel beta-sheets and an alpha-helix, followed by a long loop to the C-terminal end of the finger. The N-terminal part, which includes the helix, is similar in structure, but not sequence, to the N-terminal zinc module of the glucocorticoid receptor DNA-binding domain. The helix and the loop connecting the 2 beta-sheets interact with the major groove of the DNA, while the C-terminal tail wraps around into the minor groove. Interactions between the Znf and DNA are mainly hydrophobic, explaining the preponderance of thymines in the binding site; a large number of interactions with the phosphate backbone have also been observed. Two GATA zinc fingers are found in GATA-family transcription factors. However, there are several proteins that only contains a single copy of the domain. It is also worth noting that many GATA-type Znfs have not been experimentally demonstrated to be DNA-binding domains. Furthermore, several GATA-type Znfs have been demonstrated to act as protein-recognition domains. For example, the N-terminal Znf of GATA1 binds specifically to a zinc finger from the transcriptional coregulator FOG1 (ZFPM1).

In molecular biology, the GCM transcription factors are a family of proteins which contain a GCM motif. The GCM motif is a domain that has been identified in proteins belonging to a family of transcriptional regulators involved in fundamental developmental processes which comprise Drosophila melanogaster GCM and its mammalian homologues. In GCM transcription factors the N-terminal moiety contains a DNA-binding domain of 150 amino acids. Sequence conservation is highest in this GCM domain. In contrast, the C-terminal moiety contains one or two transactivating regions and is only poorly conserved.
In molecular biology, the WAC domain is a protein domain found on the N-terminus of WSTF protein. Its function is still unknown, but putatively thought to be involved in cell growth. The protein domain has been found to be present in both prokaryotes and eukaryotes
dClock (clk) is a gene located on the 3L chromosome of Drosophila melanogaster. Mapping and cloning of the gene indicates that it is the Drosophila homolog of the mouse gene CLOCK (mClock). The Jrk mutation disrupts the transcription cycling of per and tim and manifests dominant effects.

Sex-lethal (Sxl) is a gene found in Dipteran insects, named for its mutation phenotype in Drosophila melanogaster. It is most closely related to the ELAV/HUD subfamily of splicing factors.