
Kary Banks Mullis was an American biochemist. In recognition of his role in the invention of the polymerase chain reaction (PCR) technique, he shared the 1993 Nobel Prize in Chemistry with Michael Smith and was awarded the Japan Prize in the same year. PCR became a central technique in biochemistry and molecular biology, described by The New York Times as "highly original and significant, virtually dividing biology into the two epochs of before PCR and after PCR."
Molecular biology is a branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions.

The polymerase chain reaction (PCR) is a method widely used to make millions to billions of copies of a specific DNA sample rapidly, allowing scientists to amplify a very small sample of DNA sufficiently to enable detailed study. PCR was invented in 1983 by American biochemist Kary Mullis at Cetus Corporation; Mullis and biochemist Michael Smith, who had developed other essential ways of manipulating DNA; the creation for which they were jointly awarded the Nobel Prize in Chemistry in 1993.

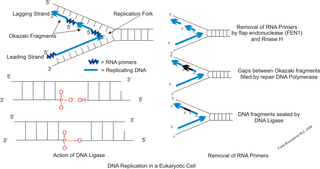
A primer is a short single-stranded nucleic acid used by all living organisms in the initiation of DNA synthesis. A synthetic primer may also be referred to as an oligo, short for oligonucleotide. DNA polymerase enzymes are only capable of adding nucleotides to the 3’-end of an existing nucleic acid, requiring a primer be bound to the template before DNA polymerase can begin a complementary strand. DNA polymerase adds nucleotides after binding to the RNA primer and synthesizes the whole strand. Later, the RNA strands must be removed accurately and replace them with DNA nucleotides forming a gap region known as a nick that is filled in using an enzyme called ligase. The removal process of the RNA primer requires several enzymes, such as Fen1, Lig1, and others that work in coordination with DNA polymerase, to ensure the removal of the RNA nucleotides and the addition of DNA nucleotides. Living organisms use solely RNA primers, while laboratory techniques in biochemistry and molecular biology that require in vitro DNA synthesis usually use DNA primers, since they are more temperature stable. Primers can be designed in laboratory for specific reactions such as polymerase chain reaction (PCR). When designing PCR primers, there are specific measures that must be taken into consideration, like the melting temperature of the primers and the annealing temperature of the reaction itself. Moreover, the DNA binding sequence of the primer in vitro has to be specifically chosen, which is done using a method called basic local alignment search tool (BLAST) that scans the DNA and finds specific and unique regions for the primer to bind.

Michael Smith was a British-born Canadian biochemist and businessman. He shared the 1993 Nobel Prize in Chemistry with Kary Mullis for his work in developing site-directed mutagenesis. Following a PhD in 1956 from the University of Manchester, he undertook postdoctoral research with Har Gobind Khorana at the British Columbia Research Council in Vancouver, British Columbia, Canada. Subsequently, Smith worked at the Fisheries Research Board of Canada Laboratory in Vancouver before being appointed a professor of biochemistry in the UBC Faculty of Medicine in 1966. Smith's career included roles as the founding director of the UBC Biotechnology Laboratory and the founding scientific leader of the Protein Engineering Network of Centres of Excellence (PENCE). In 1996 he was named Peter Wall Distinguished Professor of Biotechnology. Subsequently, he became the founding director of the Genome Sequencing Centre at the BC Cancer Research Centre.

Reverse transcription polymerase chain reaction (RT-PCR) is a laboratory technique combining reverse transcription of RNA into DNA and amplification of specific DNA targets using polymerase chain reaction (PCR). It is primarily used to measure the amount of a specific RNA. This is achieved by monitoring the amplification reaction using fluorescence, a technique called real-time PCR or quantitative PCR (qPCR). Combined RT-PCR and qPCR are routinely used for analysis of gene expression and quantification of viral RNA in research and clinical settings.

Molecular genetics is a sub-field of biology that addresses how differences in the structures or expression of DNA molecules manifests as variation among organisms. Molecular genetics often applies an "investigative approach" to determine the structure and/or function of genes in an organism's genome using genetic screens. The field of study is based on the merging of several sub-fields in biology: classical Mendelian inheritance, cellular biology, molecular biology, biochemistry, and biotechnology. Researchers search for mutations in a gene or induce mutations in a gene to link a gene sequence to a specific phenotype. Molecular genetics is a powerful methodology for linking mutations to genetic conditions that may aid the search for treatments/cures for various genetics diseases.

Har Gobind Khorana was an Indian-American biochemist. While on the faculty of the University of Wisconsin–Madison, he shared the 1968 Nobel Prize for Physiology or Medicine with Marshall W. Nirenberg and Robert W. Holley for research that showed the order of nucleotides in nucleic acids, which carry the genetic code of the cell and control the cell's synthesis of proteins. Khorana and Nirenberg were also awarded the Louisa Gross Horwitz Prize from Columbia University in the same year.

DNA synthesis is the natural or artificial creation of deoxyribonucleic acid (DNA) molecules. DNA is a macromolecule made up of nucleotide units, which are linked by covalent bonds and hydrogen bonds, in a repeating structure. DNA synthesis occurs when these nucleotide units are joined to form DNA; this can occur artificially or naturally. Nucleotide units are made up of a nitrogenous base, pentose sugar (deoxyribose) and phosphate group. Each unit is joined when a covalent bond forms between its phosphate group and the pentose sugar of the next nucleotide, forming a sugar-phosphate backbone. DNA is a complementary, double stranded structure as specific base pairing occurs naturally when hydrogen bonds form between the nucleotide bases.

DNA polymerase I is an enzyme that participates in the process of prokaryotic DNA replication. Discovered by Arthur Kornberg in 1956, it was the first known DNA polymerase. It was initially characterized in E. coli and is ubiquitous in prokaryotes. In E. coli and many other bacteria, the gene that encodes Pol I is known as polA. The E. coli Pol I enzyme is composed of 928 amino acids, and is an example of a processive enzyme — it can sequentially catalyze multiple polymerisation steps without releasing the single-stranded template. The physiological function of Pol I is mainly to support repair of damaged DNA, but it also contributes to connecting Okazaki fragments by deleting RNA primers and replacing the ribonucleotides with DNA.
In molecular biology, an amplicon is a piece of DNA or RNA that is the source and/or product of amplification or replication events. It can be formed artificially, using various methods including polymerase chain reactions (PCR) or ligase chain reactions (LCR), or naturally through gene duplication. In this context, amplification refers to the production of one or more copies of a genetic fragment or target sequence, specifically the amplicon. As it refers to the product of an amplification reaction, amplicon is used interchangeably with common laboratory terms, such as "PCR product."
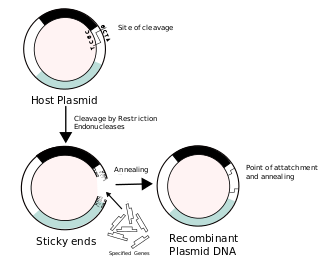
Recombinant DNA (rDNA) molecules are DNA molecules formed by laboratory methods of genetic recombination that bring together genetic material from multiple sources, creating sequences that would not otherwise be found in the genome.
Gene amplification refers to a number of natural and artificial processes by which the number of copies of a gene is increased "without a proportional increase in other genes".
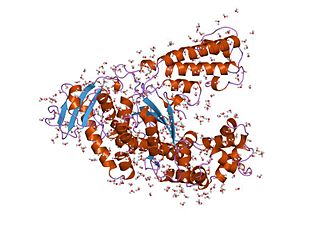
Taq polymerase is a thermostable DNA polymerase I named after the thermophilic eubacterial microorganism Thermus aquaticus, from which it was originally isolated by Chien et al. in 1976. Its name is often abbreviated to Taq or Taq pol. It is frequently used in the polymerase chain reaction (PCR), a method for greatly amplifying the quantity of short segments of DNA.
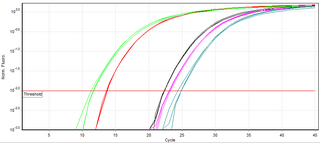
A real-time polymerase chain reaction is a laboratory technique of molecular biology based on the polymerase chain reaction (PCR). It monitors the amplification of a targeted DNA molecule during the PCR, not at its end, as in conventional PCR. Real-time PCR can be used quantitatively and semi-quantitatively.
TaqMan probes are hydrolysis probes that are designed to increase the specificity of quantitative PCR. The method was first reported in 1991 by researcher Kary Mullis at Cetus Corporation, and the technology was subsequently developed by Hoffmann-La Roche for diagnostic assays and by Applied Biosystems for research applications.
A primer binding site is a region of a nucleotide sequence where an RNA or DNA single-stranded primer binds to start replication. The primer binding site is on one of the two complementary strands of a double-stranded nucleotide polymer, in the strand which is to be copied, or is within a single-stranded nucleotide polymer sequence.

The history of the polymerase chain reaction (PCR) has variously been described as a classic "Eureka!" moment, or as an example of cooperative teamwork between disparate researchers. Following is a list of events before, during, and after its development:
The versatility of polymerase chain reaction (PCR) has led to modifications of the basic protocol being used in a large number of variant techniques designed for various purposes. This article summarizes many of the most common variations currently or formerly used in molecular biology laboratories; familiarity with the fundamental premise by which PCR works and corresponding terms and concepts is necessary for understanding these variant techniques.

Molecular diagnostics is a collection of techniques used to analyze biological markers in the genome and proteome, and how their cells express their genes as proteins, applying molecular biology to medical testing. In medicine the technique is used to diagnose and monitor disease, detect risk, and decide which therapies will work best for individual patients, and in agricultural biosecurity similarly to monitor crop- and livestock disease, estimate risk, and decide what quarantine measures must be taken.















