Related Research Articles
Mass spectrometry (MS) is an analytical technique that is used to measure the mass-to-charge ratio of ions. The results are presented as a mass spectrum, a plot of intensity as a function of the mass-to-charge ratio. Mass spectrometry is used in many different fields and is applied to pure samples as well as complex mixtures.

An ion source is a device that creates atomic and molecular ions. Ion sources are used to form ions for mass spectrometers, optical emission spectrometers, particle accelerators, ion implanters and ion engines.

Tandem mass spectrometry, also known as MS/MS or MS2, is a technique in instrumental analysis where two or more mass analyzers are coupled together using an additional reaction step to increase their abilities to analyse chemical samples. A common use of tandem MS is the analysis of biomolecules, such as proteins and peptides.

In mass spectrometry, matrix-assisted laser desorption/ionization (MALDI) is an ionization technique that uses a laser energy absorbing matrix to create ions from large molecules with minimal fragmentation. It has been applied to the analysis of biomolecules and various organic molecules, which tend to be fragile and fragment when ionized by more conventional ionization methods. It is similar in character to electrospray ionization (ESI) in that both techniques are relatively soft ways of obtaining ions of large molecules in the gas phase, though MALDI typically produces far fewer multi-charged ions.
Infrared multiple photon dissociation (IRMPD) is a technique used in mass spectrometry to fragment molecules in the gas phase usually for structural analysis of the original (parent) molecule.

Electron-capture dissociation (ECD) is a method of fragmenting gas-phase ions for structure elucidation of peptides and proteins in tandem mass spectrometry. It is one of the most widely used techniques for activation and dissociation of mass selected precursor ion in MS/MS. It involves the direct introduction of low-energy electrons to trapped gas-phase ions.

The history of mass spectrometry has its roots in physical and chemical studies regarding the nature of matter. The study of gas discharges in the mid 19th century led to the discovery of anode and cathode rays, which turned out to be positive ions and electrons. Improved capabilities in the separation of these positive ions enabled the discovery of stable isotopes of the elements. The first such discovery was with the element neon, which was shown by mass spectrometry to have at least two stable isotopes: 20Ne and 22Ne. Mass spectrometers were used in the Manhattan Project for the separation of isotopes of uranium necessary to create the atomic bomb.

Electron-transfer dissociation (ETD) is a method of fragmenting multiply-charged gaseous macromolecules in a mass spectrometer between the stages of tandem mass spectrometry (MS/MS). Similar to electron-capture dissociation, ETD induces fragmentation of large, multiply-charged cations by transferring electrons to them. ETD is used extensively with polymers and biological molecules such as proteins and peptides for sequence analysis. Transferring an electron causes peptide backbone cleavage into c- and z-ions while leaving labile post translational modifications (PTM) intact. The technique only works well for higher charge state peptide or polymer ions (z>2). However, relative to collision-induced dissociation (CID), ETD is advantageous for the fragmentation of longer peptides or even entire proteins. This makes the technique important for top-down proteomics. The method was developed by Hunt and coworkers at the University of Virginia.

Desorption electrospray ionization (DESI) is an ambient ionization technique that can be coupled to mass spectrometry (MS) for chemical analysis of samples at atmospheric conditions. Coupled ionization sources-MS systems are popular in chemical analysis because the individual capabilities of various sources combined with different MS systems allow for chemical determinations of samples. DESI employs a fast-moving charged solvent stream, at an angle relative to the sample surface, to extract analytes from the surfaces and propel the secondary ions toward the mass analyzer. This tandem technique can be used to analyze forensics analyses, pharmaceuticals, plant tissues, fruits, intact biological tissues, enzyme-substrate complexes, metabolites and polymers. Therefore, DESI-MS may be applied in a wide variety of sectors including food and drug administration, pharmaceuticals, environmental monitoring, and biotechnology.
Robert Graham Cooks is the Henry Bohn Hass Distinguished Professor of Chemistry in the Aston Laboratories for Mass Spectrometry at Purdue University. He is an ISI Highly Cited Chemist, with over 1,000 publications and an H-index of 134.

Laser spray ionization refers to one of several methods for creating ions using a laser interacting with a spray of neutral particles or ablating material to create a plume of charged particles. The ions thus formed can be separated by m/z with mass spectrometry. Laser spray is one of several ion sources that can be coupled with liquid chromatography-mass spectrometry for the detection of larger molecules.

Matrix-assisted laser desorption electrospray ionization (MALDESI) was first introduced in 2006 as a novel ambient ionization technique which combines the benefits of electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI). An infrared (IR) or ultraviolet (UV) laser can be utilized in MALDESI to resonantly excite an endogenous or exogenous matrix. The term ‘matrix’ refers to any molecule that is present in large excess and absorbs the energy of the laser, thus facilitating desorption of analyte molecules. The original MALDESI design was implemented using common organic matrices, similar to those used in MALDI, along with a UV laser. The current MALDESI source employs endogenous water or a thin layer of exogenously deposited ice as the energy-absorbing matrix where O-H symmetric and asymmetric stretching bonds are resonantly excited by a mid-IR laser.

Delayed extraction is a method used with a time-of-flight mass spectrometer in which the accelerating voltage is applied after some short time delay following pulsed laser desorption/ionization from a flat surface of target plate or, in other implementation, pulsed electron ionization or Resonance enhanced multiphoton ionization in some narrow space between two plates of the ion extraction system. The extraction delay can produce time-of-flight compensation for ion energy spread and improve mass resolution.

Ambient ionization is a form of ionization in which ions are formed in an ion source outside the mass spectrometer without sample preparation or separation. Ions can be formed by extraction into charged electrospray droplets, thermally desorbed and ionized by chemical ionization, or laser desorbed or ablated and post-ionized before they enter the mass spectrometer.

A triple quadrupole mass spectrometer (TQMS), is a tandem mass spectrometer consisting of two quadrupole mass analyzers in series, with a (non-mass-resolving) radio frequency (RF)–only quadrupole between them to act as a cell for collision-induced dissociation. This configuration is often abbreviated QqQ, here Q1q2Q3.

In mass spectrometry, fragmentation is the dissociation of energetically unstable molecular ions formed from passing the molecules in the ionization chamber of a mass spectrometer. The fragments of a molecule cause a unique pattern in the mass spectrum. These reactions are well documented over the decades and fragmentation pattern is useful to determine the molar weight and structural information of the unknown molecule. Fragmentation that occurs in tandem mass spectrometry experiments has been a recent focus of research, because this data helps facilitate the identification of molecules.

Collision-induced dissociation (CID), also known as collisionally activated dissociation (CAD), is a mass spectrometry technique to induce fragmentation of selected ions in the gas phase. The selected ions are usually accelerated by applying an electrical potential to increase the ion kinetic energy and then allowed to collide with neutral molecules. In the collision some of the kinetic energy is converted into internal energy which results in bond breakage and the fragmentation of the molecular ion into smaller fragments. These fragment ions can then be analyzed by tandem mass spectrometry.
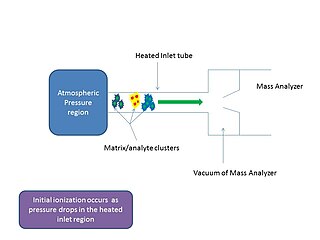
In mass spectrometry, matrix-assisted ionization is a low fragmentation (soft) ionization technique which involves the transfer of particles of the analyte and matrix sample from atmospheric pressure (AP) to the heated inlet tube connecting the AP region to the vacuum of the mass analyzer.

Gary Glish is an American analytical chemist at the University of North Carolina at Chapel Hill. He is a leading researcher in the fields of mass spectrometry, ion chemistry, and biomolecule analysis.
References
- ↑ Busch, Kenneth L. (February 2002). "SAMS:Speaking with Acronyms in Mass Spectrometry" (PDF). Spectroscopy. 17 (2). Archived from the original (PDF) on 2011-04-19. Retrieved 2009-02-02.
- ↑ Busch, Kenneth L. (May 1, 2006). "Acronyms in Mass Spectrometry". spectroscopyonline.com. Retrieved September 22, 2015.
- ↑ He, Jiuming; Tang, Fei; Luo, Zhigang; Chen, Yi; Xu, Jing; Zhang, Ruiping; Wang, Xiaohao; Abliz, Zeper (2011). "Air flow assisted ionization for remote sampling of ambient mass spectrometry and its application". Rapid Communications in Mass Spectrometry. 25 (7): 843–850. doi:10.1002/rcm.4920. ISSN 0951-4198. PMID 21416520.
- ↑ Peterson, Lowell (1962). "Mass Spectrometer All-Glass Heated Inlet". Analytical Chemistry. 34 (13): 1850–1851. doi:10.1021/ac60193a054. ISSN 0003-2700.
- ↑ O’Brien, Jeremy T.; Williams, Evan R.; Holman, Hoi-Ying N. (2015). "Ambient Infrared Laser Ablation Mass Spectrometry (AIRLAB-MS) of Live Plant Tissue with Plume Capture by Continuous Flow Solvent Probe". Analytical Chemistry. 87 (5): 2631–2638. doi:10.1021/ac503383p. ISSN 0003-2700. PMID 25622206.
- ↑ Dunbar, Robert C. (2004). "BIRD (blackbody infrared radiative dissociation): Evolution, principles, and applications". Mass Spectrometry Reviews. 23 (2): 127–58. Bibcode:2004MSRv...23..127D. doi:10.1002/mas.10074. PMID 14732935.
- ↑ Abramson, Fred P. (1994). "CRIMS: Chemical reaction interface mass spectrometry". Mass Spectrometry Reviews. 13 (4): 341–356. Bibcode:1994MSRv...13..341A. doi:10.1002/mas.1280130403.
- ↑ Hoffmann WD, Jackson GP (November 2014). "Charge transfer dissociation (CTD) mass spectrometry of peptide cations using kiloelectronvolt helium cations". Journal of the American Society for Mass Spectrometry. 25 (11): 1939–43. Bibcode:2014JASMS..25.1939H. doi:10.1007/s13361-014-0989-6. PMID 25231159.
- ↑ Buriak, Jillian M.; Wei, Jing; Siuzdak, Gary (1999). "Desorption–ionization mass spectrometry on porous silicon". Nature. 399 (6733): 243–6. Bibcode:1999Natur.399..243W. doi:10.1038/20400. PMID 10353246.