Biography
Levine was born in West Hollywood and raised in Los Angeles. [3] Levine studied biology as an undergraduate at UC Berkeley, studying biology with Allan Wilson [3] and graduating in 1976. [4] He went on to graduate studies at Yale, where he studied with Alan Garen and in 1981 received a Ph.D. in molecular biophysics and biochemistry. [4]
Levine joined the Princeton faculty in 2015, and had been a professor at UC Berkeley after leaving UCSD in 1996. [5]
Discoveries
Homeobox discovery
Levine was a post-doc with Walter Gehring in Switzerland from 1982 to 1983. [6] There, he co-discovered the homeobox with Ernst Hafen and fellow post-doc William McGinnis: [7]
After learning that Ultrabithorax, a gene that specifies the development of wings, showed a localized pattern of expression similar to that of Antennapedia, they decided to revisit the classic papers of Ed Lewis. In 1978, Lewis had proposed that all these homeotic genes (the ones that tell animals where to put a wing and where to put a leg and so on) arose from a common ancestral gene. So McGinnis carved up the Antennapedia gene and, using those pieces as probes, the trio identified eight genes, which turned out to be the eight homeotic genes in flies. "That pissed off a lot of people," says Levine. "The homeotic genes were the trophies of the Drosophila genome. And we got 'em all. I mean, we got 'em all!" Far from being humble, Levine says, "We were like, 'We kicked your ass pretty good, didn't we, baby!' Those were the days." [3]
Discovery of the eve stripe 2 enhancer
Levine briefly returned to UC Berkeley as a postdoctoral fellow [4] with Gerry Rubin. [8] He then joined the faculty of Columbia University, where he "led the discovery of the modular organization of the regulatory regions of developmental genes." [9] After isolating the even-skipped (eve) gene, Levine's team determined that each of the seven stripes was produced by separate enhancers. [3] With further study they discovered that both a set of activators and a set of repressors worked together to shape the expression of eve in the second stripe, and determined that the repressors shut down only their binding enhancers, leaving other enhancers free of repression. [3] Joseph Corbo said of the work,
"Before Levine's studies of even-skipped stripe 2, it wasn't clear how you generated spatially restricted patterns of gene expression from initially broad crude gradients of morphogens. I think that the even-skipped stripe 2 studies were the defining studies that showed how an organism can interpret those gradients and turn them into specific patterns of gene expression. To me that's Mike's crowning achievement." [3]
Discoveries in the ascidian Ciona
After earning tenure in only four years at Columbia, [3] Levine moved to UCSD in 1991, [4] where he added the sea squirt, Ciona intestinalis , to his repertoire. Although much of Levine's work, including his homeobox studies, has been done in Drosophila [6] Levine's team is also prominent in work with the sea squirt, Ciona intestinalis , an invertebrate that facilitates study of development. [3] For example, this work included insights into classical myodeterminants [10] [11] [12] and the composition of the notochord, the defining tissue of the chordate phylum. [13]
Professional relations
Levine cites as a significant influence his instructor Fred Wilt (taking his developmental biology class "was probably the single most galvanizing experience I had in terms of defining my future goals"), [8] and cites fellow scientists Eric Davidson, Peter Lawrence and Christiane Nusslein-Volhard as "mentors [and] friends ... over the years". [8]
On choosing to become a research biologist, he described some family pressure to become a doctor ("Coming from a modest background, particularly a Jewish family, the pressure to become a doctor was intense"), [3]
Fellow biologist Sean Carroll said of Levine, "Mike's work has done for animal development what the work on the lac operon and phage lambda did for understanding gene regulation in simpler organisms ... [Those] two big discoveries had a very large conceptual significance for developmental biology and by extension for evolutionary biology." [9]

Evolutionary developmental biology is a field of biological research that compares the developmental processes of different organisms to infer how developmental processes evolved.

A homeobox is a DNA sequence, around 180 base pairs long, that regulates large-scale anatomical features in the early stages of embryonic development. Mutations in a homeobox may change large-scale anatomical features of the full-grown organism.

Drosophila embryogenesis, the process by which Drosophila embryos form, is a favorite model system for genetics and developmental biology. The study of its embryogenesis unlocked the century-long puzzle of how development was controlled, creating the field of evolutionary developmental biology. The small size, short generation time, and large brood size make it ideal for genetic studies. Transparent embryos facilitate developmental studies. Drosophila melanogaster was introduced into the field of genetic experiments by Thomas Hunt Morgan in 1909.

In evolutionary developmental biology, homeosis is the transformation of one organ into another, arising from mutation in or misexpression of certain developmentally critical genes, specifically homeotic genes. In animals, these developmental genes specifically control the development of organs on their anteroposterior axis. In plants, however, the developmental genes affected by homeosis may control anything from the development of a stamen or petals to the development of chlorophyll. Homeosis may be caused by mutations in Hox genes, found in animals, or others such as the MADS-box family in plants. Homeosis is a characteristic that has helped insects become as successful and diverse as they are.
William McGinnis, Ph.D. is a molecular biologist and professor of biology at the University of California San Diego. At UC San Diego he has also served as the Chairman of the Department of Biology from July 1998 - June 1999, as Associate Dean of the Division of Natural Sciences from July 1, 1999 - June 2000, and as Interim Dean of the newly established Division of Biological Sciences from July 1, 2000 - February 1, 2001. Dr. McGinnis was appointed Dean of the Divisional Biological Sciences on July 1, 2013.

A morphogen is a substance whose non-uniform distribution governs the pattern of tissue development in the process of morphogenesis or pattern formation, one of the core processes of developmental biology, establishing positions of the various specialized cell types within a tissue. More specifically, a morphogen is a signaling molecule that acts directly on cells to produce specific cellular responses depending on its local concentration.
Hox genes, a subset of homeobox genes, are a group of related genes that specify regions of the body plan of an embryo along the head-tail axis of animals. Hox proteins encode and specify the characteristics of 'position', ensuring that the correct structures form in the correct places of the body. For example, Hox genes in insects specify which appendages form on a segment, and Hox genes in vertebrates specify the types and shape of vertebrae that will form. In segmented animals, Hox proteins thus confer segmental or positional identity, but do not form the actual segments themselves.

Walter Jakob Gehring was a Swiss developmental biologist who was a professor at the Biozentrum Basel of the University of Basel, Switzerland. He obtained his PhD at the University of Zurich in 1965 and after two years as a research assistant of Ernst Hadorn he joined Alan Garen's group at Yale University in New Haven as a postdoctoral fellow.

A gap gene is a type of gene involved in the development of the segmented embryos of some arthropods. Gap genes are defined by the effect of a mutation in that gene, which causes the loss of contiguous body segments, resembling a gap in the normal body plan. Each gap gene, therefore, is necessary for the development of a section of the organism.

Homeobox protein Nkx-2.5 is a protein that in humans is encoded by the NKX2-5 gene.

Homeobox protein Hox-B5 is a protein that in humans is encoded by the HOXB5 gene.
Homeotic genes are genes which regulate the development of anatomical structures in various organisms such as echinoderms, insects, mammals, and plants. Homeotic genes often encode transcription factor proteins, and these proteins affect development by regulating downstream gene networks involved in body patterning.
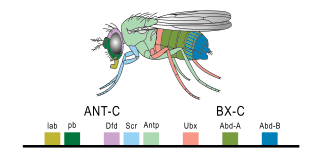
The Bithorax complex (BX-C) is one of two Drosophila melanogaster homeotic gene complexes, located on the right arm of chromosome 3. It is responsible for the differentiation of the posterior two-thirds of the fly by the regulation of three genes within the complex: Ultrabithorax (Ubx), abdominal A (abd-A), and Abdominal B (Abd-B).
Homeotic selector genes confer segment identity in Drosophila. They encode homeodomain proteins which interact with Hox and other homeotic genes to initiate segment-specific gene regulation. Homeodomain proteins are transcription factors that share a DNA-binding domain called the homeodomain. Changes in the expression and function of homeotic genes are responsible for the changes in the morphology of the limbs of arthropods as well as in the axial skeletons of vertebrates. Mutations in homeotic selector genes do not lead to elimination of a segment or pattern, but instead cause the segment to develop incorrectly.
Zerknüllt is a gene in the Antennapedia complex of Drosophila and other insects, where it operates very differently from the canonical Hox genes in the same gene cluster. Comparison of Hox genes between species showed that the Zerknüllt gene evolved from one of the standard Hox genes in insects through accumulating many amino acid changes, changing expression pattern, losing ancestral function and gaining a new function.
Albert Erives is a developmental geneticist who studies transcriptional enhancers underlying animal development and diseases of development (cancers). Erives also proposed the pacRNA model for the dual origin of the genetic code and universal homochirality. He is known for work at the intersection of genetics, evolution, developmental biology, and gene regulation. He has worked at the California Institute of Technology, University of California, Berkeley, and Dartmouth College, and is an associate professor at the University of Iowa.
Ueli Schibler is a Swiss biologist, chronobiologist and a professor at the University of Geneva. His research has contributed significantly to the field of chronobiology and the understanding of circadian clocks in the body. Several of his studies have demonstrated strong evidence for the existence of robust, self-sustaining circadian clocks in the peripheral tissues.
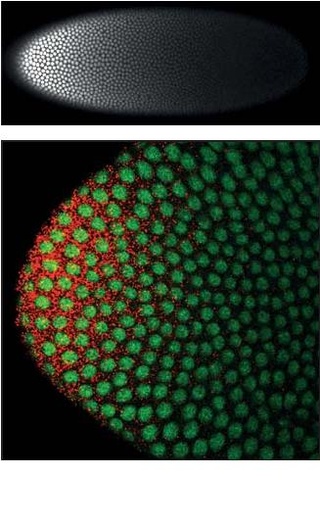
Homeotic protein bicoid is encoded by the bcd maternal effect gene in Drosophilia. Homeotic protein bicoid concentration gradient patterns the anterior-posterior (A-P) axis during Drosophila embryogenesis. Bicoid was the first protein demonstrated to act as a morphogen. Although bicoid is important for the development of Drosophila and other higher dipterans, it is absent from most other insects, where its role is accomplished by other genes.
Thomas Charles Kaufman is an American geneticist. He is known for his work on the zeste-white region of the Drosophila X chromosome. He is currently a Distinguished Professor of biology at Indiana University, where he conducts his current research on Homeotic Genes in evolution and development.
Shadow enhancers are groups of two or more enhancers that control the same target gene and drive overlapping spatiotemporal expression patterns. Shadow enhancers are found in a wide range of organisms, from insects to plants to mammals, particularly in association with developmental genes. While seemingly redundant, the individual enhancers of a shadow enhancer group have been shown to be critical for proper gene expression in the face of both environmental and genetic perturbations. Such perturbations may exacerbate fluctuations in upstream regulators.










