
An axon, or nerve fiber, is a long, slender projection of a nerve cell, or neuron, in vertebrates, that typically conducts electrical impulses known as action potentials away from the nerve cell body. The function of the axon is to transmit information to different neurons, muscles, and glands. In certain sensory neurons, such as those for touch and warmth, the axons are called afferent nerve fibers and the electrical impulse travels along these from the periphery to the cell body and from the cell body to the spinal cord along another branch of the same axon. Axon dysfunction can be the cause of many inherited and acquired neurological disorders that affect both the peripheral and central neurons. Nerve fibers are classed into three types – group A nerve fibers, group B nerve fibers, and group C nerve fibers. Groups A and B are myelinated, and group C are unmyelinated. These groups include both sensory fibers and motor fibers. Another classification groups only the sensory fibers as Type I, Type II, Type III, and Type IV.
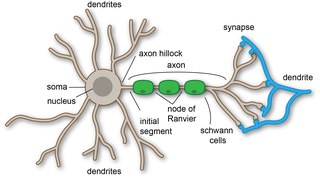
A dendrite or dendron is a branched protoplasmic extension of a nerve cell that propagates the electrochemical stimulation received from other neural cells to the cell body, or soma, of the neuron from which the dendrites project. Electrical stimulation is transmitted onto dendrites by upstream neurons via synapses which are located at various points throughout the dendritic tree.

Within a nervous system, a neuron, neurone, or nerve cell is an electrically excitable cell that fires electric signals called action potentials across a neural network. Neurons communicate with other cells via synapses - specialized connections that commonly use minute amounts of chemical neurotransmitters to pass the electric signal from the presynaptic neuron to the target cell through the synaptic gap. The neuron is the main component of nervous tissue in all animals except sponges and placozoa. Non-animals like plants and fungi do not have nerve cells.
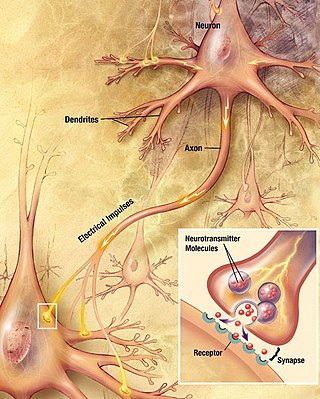
Chemical synapses are biological junctions through which neurons' signals can be sent to each other and to non-neuronal cells such as those in muscles or glands. Chemical synapses allow neurons to form circuits within the central nervous system. They are crucial to the biological computations that underlie perception and thought. They allow the nervous system to connect to and control other systems of the body.

An action potential occurs when the membrane potential of a specific cell rapidly rises and falls. This depolarization then causes adjacent locations to similarly depolarize. Action potentials occur in several types of animal cells, called excitable cells, which include neurons, muscle cells, and in some plant cells. Certain endocrine cells such as pancreatic beta cells, and certain cells of the anterior pituitary gland are also excitable cells.
Nervous tissue, also called neural tissue, is the main tissue component of the nervous system. The nervous system regulates and controls body functions and activity. It consists of two parts: the central nervous system (CNS) comprising the brain and spinal cord, and the peripheral nervous system (PNS) comprising the branching peripheral nerves. It is composed of neurons, also known as nerve cells, which receive and transmit impulses, and neuroglia, also known as glial cells or glia, which assist the propagation of the nerve impulse as well as provide nutrients to the neurons.

Pyramidal cells, or pyramidal neurons, are a type of multipolar neuron found in areas of the brain including the cerebral cortex, the hippocampus, and the amygdala. Pyramidal cells are the primary excitation units of the mammalian prefrontal cortex and the corticospinal tract. Pyramidal neurons are also one of two cell types where the characteristic sign, Negri bodies, are found in post-mortem rabies infection. Pyramidal neurons were first discovered and studied by Santiago Ramón y Cajal. Since then, studies on pyramidal neurons have focused on topics ranging from neuroplasticity to cognition.

In cellular neuroscience, the soma, perikaryon, neurocyton, or cell body is the bulbous, non-process portion of a neuron or other brain cell type, containing the cell nucleus. Although it is often used to refer to neurons, it can also refer to other cell types as well, including astrocytes, oligodendrocytes, and microglia. There are many different specialized types of neurons, and their sizes vary from as small as about 5 micrometres to over 10 millimetres for some of the smallest and largest neurons of invertebrates, respectively.

In the nervous system, a synapse is a structure that permits a neuron to pass an electrical or chemical signal to another neuron or to the target effector cell.
GENESIS is a simulation environment for constructing realistic models of neurobiological systems at many levels of scale including: sub-cellular processes, individual neurons, networks of neurons, and neuronal systems. These simulations are “computer-based implementations of models whose primary objective is to capture what is known of the anatomical structure and physiological characteristics of the neural system of interest”. GENESIS is intended to quantify the physical framework of the nervous system in a way that allows for easy understanding of the physical structure of the nerves in question. “At present only GENESIS allows parallelized modeling of single neurons and networks on multiple-instruction-multiple-data parallel computers.” Development of GENESIS software spread from its home at Caltech to labs at the University of Texas at San Antonio, the University of Antwerp, the National Centre for Biological Sciences in Bangalore, the University of Colorado, the Pittsburgh Supercomputing Center, the San Diego Supercomputer Center, and Emory University.

The Calyx of Held is a particularly large synapse in the mammalian auditory central nervous system, so named after Hans Held who first described it in his 1893 article Die centrale Gehörleitung because of its resemblance to the calyx of a flower. Globular bushy cells in the anteroventral cochlear nucleus (AVCN) send axons to the contralateral medial nucleus of the trapezoid body (MNTB), where they synapse via these calyces on MNTB principal cells. These principal cells then project to the ipsilateral lateral superior olive (LSO), where they inhibit postsynaptic neurons and provide a basis for interaural level detection (ILD), required for high frequency sound localization. This synapse has been described as the largest in the brain.
Neural backpropagation is the phenomenon in which, after the action potential of a neuron creates a voltage spike down the axon, another impulse is generated from the soma and propagates towards the apical portions of the dendritic arbor or dendrites. In addition to active backpropagation of the action potential, there is also passive electrotonic spread. While there is ample evidence to prove the existence of backpropagating action potentials, the function of such action potentials and the extent to which they invade the most distal dendrites remain highly controversial.

Biological neuron models, also known as a spiking neuron models, are mathematical descriptions of the properties of certain cells in the nervous system that generate sharp electrical potentials across their cell membrane, roughly one millisecond in duration, called action potentials or spikes. Since spikes are transmitted along the axon and synapses from the sending neuron to many other neurons, spiking neurons are considered to be a major information processing unit of the nervous system. Spiking neuron models can be divided into different categories: the most detailed mathematical models are biophysical neuron models that describe the membrane voltage as a function of the input current and the activation of ion channels. Mathematically simpler are integrate-and-fire models that describe the membrane voltage as a function of the input current and predict the spike times without a description of the biophysical processes that shape the time course of an action potential. Even more abstract models only predict output spikes as a function of the stimulation where the stimulation can occur through sensory input or pharmacologically. This article provides a short overview of different spiking neuron models and links, whenever possible to experimental phenomena. It includes deterministic and probabilistic models.

In neurophysiology, a dendritic spike refers to an action potential generated in the dendrite of a neuron. Dendrites are branched extensions of a neuron. They receive electrical signals emitted from projecting neurons and transfer these signals to the cell body, or soma. Dendritic signaling has traditionally been viewed as a passive mode of electrical signaling. Unlike its axon counterpart which can generate signals through action potentials, dendrites were believed to only have the ability to propagate electrical signals by physical means: changes in conductance, length, cross sectional area, etc. However, the existence of dendritic spikes was proposed and demonstrated by W. Alden Spencer, Eric Kandel, Rodolfo Llinás and coworkers in the 1960s and a large body of evidence now makes it clear that dendrites are active neuronal structures. Dendrites contain voltage-gated ion channels giving them the ability to generate action potentials. Dendritic spikes have been recorded in numerous types of neurons in the brain and are thought to have great implications in neuronal communication, memory, and learning. They are one of the major factors in long-term potentiation.

Axon terminals are distal terminations of the telodendria (branches) of an axon. An axon, also called a nerve fiber, is a long, slender projection of a nerve cell, or neuron, that conducts electrical impulses called action potentials away from the neuron's cell body, or soma, in order to transmit those impulses to other neurons, muscle cells or glands.
Cellular neuroscience is a branch of neuroscience concerned with the study of neurons at a cellular level. This includes morphology and physiological properties of single neurons. Several techniques such as intracellular recording, patch-clamp, and voltage-clamp technique, pharmacology, confocal imaging, molecular biology, two photon laser scanning microscopy and Ca2+ imaging have been used to study activity at the cellular level. Cellular neuroscience examines the various types of neurons, the functions of different neurons, the influence of neurons upon each other, and how neurons work together.

Nonsynaptic plasticity is a form of neuroplasticity that involves modification of ion channel function in the axon, dendrites, and cell body that results in specific changes in the integration of excitatory postsynaptic potentials and inhibitory postsynaptic potentials. Nonsynaptic plasticity is a modification of the intrinsic excitability of the neuron. It interacts with synaptic plasticity, but it is considered a separate entity from synaptic plasticity. Intrinsic modification of the electrical properties of neurons plays a role in many aspects of plasticity from homeostatic plasticity to learning and memory itself. Nonsynaptic plasticity affects synaptic integration, subthreshold propagation, spike generation, and other fundamental mechanisms of neurons at the cellular level. These individual neuronal alterations can result in changes in higher brain function, especially learning and memory. However, as an emerging field in neuroscience, much of the knowledge about nonsynaptic plasticity is uncertain and still requires further investigation to better define its role in brain function and behavior.
Network of human nervous system comprises nodes that are connected by links. The connectivity may be viewed anatomically, functionally, or electrophysiologically. These are presented in several Wikipedia articles that include Connectionism, Biological neural network, Artificial neural network, Computational neuroscience, as well as in several books by Ascoli, G. A. (2002), Sterratt, D., Graham, B., Gillies, A., & Willshaw, D. (2011), Gerstner, W., & Kistler, W. (2002), and Rumelhart, J. L., McClelland, J. L., and PDP Research Group (1986) among others. The focus of this article is a comprehensive view of modeling a neural network. Once an approach based on the perspective and connectivity is chosen, the models are developed at microscopic, mesoscopic, or macroscopic (system) levels. Computational modeling refers to models that are developed using computing tools.
Compartmental modelling of dendrites deals with multi-compartment modelling of the dendrites, to make the understanding of the electrical behavior of complex dendrites easier. Basically, compartmental modelling of dendrites is a very helpful tool to develop new biological neuron models. Dendrites are very important because they occupy the most membrane area in many of the neurons and give the neuron an ability to connect to thousands of other cells. Originally the dendrites were thought to have constant conductance and current but now it has been understood that they may have active Voltage-gated ion channels, which influences the firing properties of the neuron and also the response of neuron to synaptic inputs. Many mathematical models have been developed to understand the electric behavior of the dendrites. Dendrites tend to be very branchy and complex, so the compartmental approach to understand the electrical behavior of the dendrites makes it very useful.
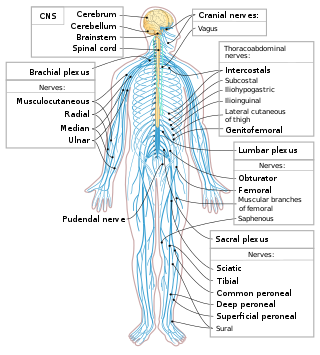
The following Diagram is provided as an overview of and topical guide to the human nervous system:















