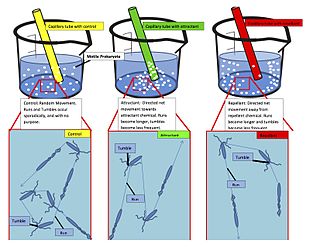
Chemotaxis is the movement of an organism or entity in response to a chemical stimulus. Somatic cells, bacteria, and other single-cell or multicellular organisms direct their movements according to certain chemicals in their environment. This is important for bacteria to find food by swimming toward the highest concentration of food molecules, or to flee from poisons. In multicellular organisms, chemotaxis is critical to early development and development as well as in normal function and health. In addition, it has been recognized that mechanisms that allow chemotaxis in animals can be subverted during cancer metastasis. The aberrant chemotaxis of leukocytes and lymphocytes also contribute to inflammatory diseases such as atherosclerosis, asthma, and arthritis. Sub-cellular components, such as the polarity patch generated by mating yeast, may also display chemotactic behavior.

A pilus is a hair-like appendage found on the surface of many bacteria and archaea. The terms pilus and fimbria can be used interchangeably, although some researchers reserve the term pilus for the appendage required for bacterial conjugation. All conjugative pili are primarily composed of pilin – fibrous proteins, which are oligomeric.

A flagellum is a hairlike appendage that protrudes from certain plant and animal sperm cells, and from a wide range of microorganisms to provide motility. Many protists with flagella are termed as flagellates.

The cilium, is a membrane-bound organelle found on most types of eukaryotic cell. Cilia are absent in bacteria and archaea. The cilium has the shape of a slender threadlike projection that extends from the surface of the much larger cell body. Eukaryotic flagella found on sperm cells and many protozoans have a similar structure to motile cilia that enables swimming through liquids; they are longer than cilia and have a different undulating motion.

Enterococcus is a large genus of lactic acid bacteria of the phylum Bacillota. Enterococci are gram-positive cocci that often occur in pairs (diplococci) or short chains, and are difficult to distinguish from streptococci on physical characteristics alone. Two species are common commensal organisms in the intestines of humans: E. faecalis (90–95%) and E. faecium (5–10%). Rare clusters of infections occur with other species, including E. casseliflavus, E. gallinarum, and E. raffinosus.

Vibrio is a genus of Gram-negative bacteria, possessing a curved-rod (comma) shape, several species of which can cause foodborne infection, usually associated with eating undercooked seafood. Being highly salt tolerant and unable to survive in fresh water, Vibrio spp. are commonly found in various salt water environments. Vibrio spp. are facultative anaerobes that test positive for oxidase and do not form spores. All members of the genus are motile. They are able to have polar or lateral flagellum with or without sheaths. Vibrio species typically possess two chromosomes, which is unusual for bacteria. Each chromosome has a distinct and independent origin of replication, and are conserved together over time in the genus. Recent phylogenies have been constructed based on a suite of genes.

Proteus mirabilis is a Gram-negative, facultatively anaerobic, rod-shaped bacterium. It shows swarming motility and urease activity. P. mirabilis causes 90% of all Proteus infections in humans. It is widely distributed in soil and water. Proteus mirabilis can migrate across the surface of solid media or devices using a type of cooperative group motility called swarming. Proteus mirabilis is most frequently associated with infections of the urinary tract, especially in complicated or catheter-associated urinary tract infections.

Enterococcus faecalis – formerly classified as part of the group D Streptococcus system – is a Gram-positive, commensal bacterium inhabiting the gastrointestinal tracts of humans. Like other species in the genus Enterococcus, E. faecalis is found in healthy humans and can be used as a probiotic. The probiotic strains such as Symbioflor1 and EF-2001 are characterized by the lack of specific genes related to drug resistance and pathogenesis. As an opportunistic pathogen, E. faecalis can cause life-threatening infections, especially in the nosocomial (hospital) environment, where the naturally high levels of antibiotic resistance found in E. faecalis contribute to its pathogenicity. E. faecalis has been frequently found in reinfected, root canal-treated teeth in prevalence values ranging from 30% to 90% of the cases. Re-infected root canal-treated teeth are about nine times more likely to harbor E. faecalis than cases of primary infections.
Members of the genus Selenomonas are referred to trivially as selenomonads. The genus Selenomonas constitutes a group of motile crescent-shaped bacteria and includes species living in the gastrointestinal tracts of animals, in particular the ruminants. A number of smaller forms discovered with the light microscope are now in culture but many, especially the large selenomonads are not, owing to their fastidious and incompletely known growth requirements.

Bile Esculin Agar (BEA) is a selective differential agar used to isolate and identify members of the genus Enterococcus, formerly part of the "group D streptococci".
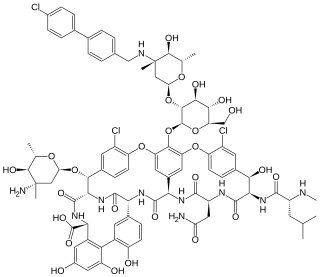
Oritavancin, sold under the brand name Orbactiv among others, is a semisynthetic glycopeptide antibiotic medication for the treatment of serious Gram-positive bacterial infections. Its chemical structure as a lipoglycopeptide is similar to vancomycin.
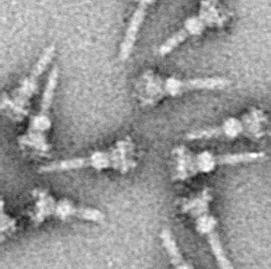
The type III secretion system, also called the injectisome, is one of the bacterial secretion systems used by bacteria to secrete their effector proteins into the host's cells to promote virulence and colonisation. The T3SS is a needle-like protein complex found in several species of pathogenic gram-negative bacteria.

Swarming motility is a rapid and coordinated translocation of a bacterial population across solid or semi-solid surfaces, and is an example of bacterial multicellularity and swarm behaviour. Swarming motility was first reported by Jorgen Henrichsen and has been mostly studied in genus Serratia, Salmonella, Aeromonas, Bacillus, Yersinia, Pseudomonas, Proteus, Vibrio and Escherichia.

Bacterial motility is the ability of bacteria to move independently using metabolic energy. Most motility mechanisms which evolved among bacteria also evolved in parallel among the archaea. Most rod-shaped bacteria can move using their own power, which allows colonization of new environments and discovery of new resources for survival. Bacterial movement depends not only on the characteristics of the medium, but also on the use of different appendages to propel. Swarming and swimming movements are both powered by rotating flagella. Whereas swarming is a multicellular 2D movement over a surface and requires the presence of surfactants, swimming is movement of individual cells in liquid environments.

Gliding motility is a type of translocation used by microorganisms that is independent of propulsive structures such as flagella, pili, and fimbriae. Gliding allows microorganisms to travel along the surface of low aqueous films. The mechanisms of this motility are only partially known.
Bacterial morphological plasticity refers to changes in the shape and size that bacterial cells undergo when they encounter stressful environments. Although bacteria have evolved complex molecular strategies to maintain their shape, many are able to alter their shape as a survival strategy in response to protist predators, antibiotics, the immune response, and other threats.
The archaellum is a unique structure on the cell surface of many archaea, that allows for swimming motility. The archaellum consists of a rigid helical filament that is attached to the cell membrane by a molecular motor. This molecular motor – composed of cytosolic, membrane, and pseudo-periplasmic proteins – is responsible for the assembly of the filament and, once assembled, for its rotation. The rotation of the filament propels archaeal cells in liquid medium, in a manner similar to the propeller of a boat. The bacterial analog of the archaellum is the flagellum, which is also responsible for their swimming motility and can also be compared to a rotating corkscrew. Although the movement of archaella and flagella is sometimes described as "whip-like", this is incorrect, as only cilia from Eukaryotes move in this manner. Indeed, even "flagellum" is a misnomer, as bacterial flagella also work as propeller-like structures.
Enterococcus raffinosus is a bacterial species of the Gram-positive genus Enterococcus, named for its facultative anaerobic metabolism, including the ability to ferment the trisaccharide raffinose. This mesophilic microaerophile has optimal growth at 37°C in Columbia Blood Medium. It has an ovoid morphology categorized as coccal with arrangement singly, in pairs, or short chains.

Social motility describes the motile movement of groups of cells that communicate with each other to coordinate movement based on external stimuli. There are multiple varieties of each kingdom that express social motility that provides a unique evolutionary advantages that other species do not possess. This has made them lethal killers such as African trypanosomiasis, or Myxobacteria. These evolutionary advantages have proven to increase survival rate among socially motile bacteria whether it be the ability to evade predators or communication within a swarm to form spores for long term hibernation in times of low nutrients or toxic environments.

Marine prokaryotes are marine bacteria and marine archaea. They are defined by their habitat as prokaryotes that live in marine environments, that is, in the saltwater of seas or oceans or the brackish water of coastal estuaries. All cellular life forms can be divided into prokaryotes and eukaryotes. Eukaryotes are organisms whose cells have a nucleus enclosed within membranes, whereas prokaryotes are the organisms that do not have a nucleus enclosed within a membrane. The three-domain system of classifying life adds another division: the prokaryotes are divided into two domains of life, the microscopic bacteria and the microscopic archaea, while everything else, the eukaryotes, become the third domain.

















