Function
The pyruvate dehydrogenase (PDH) complex must be tightly regulated due to its central role in general metabolism. Within the complex, there are three serine residues on the E1 component that are sites for phosphorylation; this phosphorylation inactivates the complex. In humans, there have been four isozymes of pyruvate dehydrogenase kinase that have been shown to phosphorylate these three sites: PDK1, PDK2, PDK3, and PDK4. PDK4 does not incorporate the most phosphate groups per catalytic event, because it can only phosphorylate site 1 and site 2; its rate of phosphorylation is less than PDK1, equal to PDK3, and more than PDK2. When the thiamine pyrophosphate (TPP) coenzyme is bound, the rates of phosphorylation by all four isozymes are drastically affected. Site 1 is the most affected, with the rate being significantly decreased. However, overall activity by PDK4 is not affected. [9]
Regulation
As the primary regulators of a crucial step in the central metabolic pathway, the pyruvate dehydrogenase family is tightly regulated itself by a myriad of factors including transcription factors Sp1 and CCAAT box binding factor (CBF). Retinoic acid enhances PDK4 transcription by enabling retinoic acid receptor family members to recruit transcriptional coactivators to retinoic acid response elements (RAREs) in the PDK4 promoter. Transcription is also increased by inhibiting inhibitory histone deacetylases (HDACs) using trichostatin A (TSA). [10] Rosiglitazone, a thiazolidinedione known to activate the glycerol biogenesis pathway, increases PDK4 mRNA transcription in white adipose tissue, but not in liver or muscle tissue. [11] Farnesoid X receptor, or FXR, suppresses glycolysis and enhances fatty acid oxidation by increasing PDK4 expression and inactivating the PDH complex. [12] Other factors, such as insulin, directly downregulate both PDK2 and PDK4 mRNA transcription. This is done through a proposed phosphatidylinositol 3-kinase (PI3K)-dependent pathway. In fact, even when cells are exposed to dexamethasone to increase mRNA expression, insulin blocks this effect. [13] Peroxisome proliferator-activated receptors also regulate expression; PPAR alpha and delta were found to upregulate PDK4 mRNA, but PPAR gamma activation downregulated expression. [14]
Clinical significance
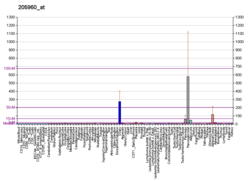
PDK4 is relevant in a variety of clinical conditions. Short-term fasting induces an increase in PDK4 transcription by about 10-fold. [15] Upon refeeding, transcription of PDK4 increased further, a surprising outlook, by about 50-fold over levels before fasting began. [16] This effect can be seen long term as well. PDK4 is overexpressed in skeletal muscle in type 2 diabetes, resulting in impaired glucose utilization. [17] In post-obese patients, there is a significant decrease in PDK4 mRNA expression, in conjunction with increased glucose uptake; this is likely due to the downregulation of PDK4 by insulin. This corroborates the concept that a lowered availability of free fatty acids affects glucose metabolism by PDH complex regulation. [18] In fact, it has been shown that insufficient downregulation of PDK mRNA in insulin-resistant individuals could be a cause of increased PDK expression leading to impaired glucose oxidation followed by increased fatty acid oxidation. [19]
Exercise has been shown to induce changes in this gene as well, and that transient changes can have a cumulative effect across many exercise sessions. The mRNA of PDK4, along with PPARGC1A , increases in both types of muscle tissue after exercise. [20] [21]
These metabolic effects can be seen in other conditions. Hypoxia is shown to induce PDK4 gene expression through the ERR gamma mechanism. [22] Conversely, PDK4 is downregulated in cardiac muscle tissue during heart failure. [23]
Cancer
The ubiquitous role of this gene lends itself to being involved in a variety of disease pathologies, including cancer. One metabolite, butyrate, induces hyperacetylation of the histones around the PDK4 gene. This is associated with a greater transcription level of PDK4 mRNA, thereby reversing the downregulation of PDK4 in colon carcinoma cells. In human colon cancer cells, targeting and inactivating the PDH complex limits the metabolic rate and regulates glutamine metabolism, thereby partially inhibiting cell growth. [24] However, PDK4 has also been shown to promote tumorigenesis and proliferation through a different pathway, the CREB-RHEB-mTORC1 signaling cascade. [25]
This page is based on this
Wikipedia article Text is available under the
CC BY-SA 4.0 license; additional terms may apply.
Images, videos and audio are available under their respective licenses.