
The major histocompatibility complex (MHC) is a large locus on vertebrate DNA containing a set of closely linked polymorphic genes that code for cell surface proteins essential for the adaptive immune system. These cell surface proteins are called MHC molecules.

The human leukocyte antigen (HLA) system or complex is a complex of genes on chromosome 6 in humans which encode cell-surface proteins responsible for regulation of the immune system. The HLA system is also known as the human version of the major histocompatibility complex (MHC) found in many animals.

Transplant rejection occurs when transplanted tissue is rejected by the recipient's immune system, which destroys the transplanted tissue. Transplant rejection can be lessened by determining the molecular similitude between donor and recipient and by use of immunosuppressant drugs after transplant.

Graft-versus-host disease (GvHD) is a syndrome, characterized by inflammation in different organs. GvHD is commonly associated with bone marrow transplants and stem cell transplants.
Alloimmunity is an immune response to nonself antigens from members of the same species, which are called alloantigens or isoantigens. Two major types of alloantigens are blood group antigens and histocompatibility antigens. In alloimmunity, the body creates antibodies against the alloantigens, attacking transfused blood, allotransplanted tissue, and even the fetus in some cases. Alloimmune (isoimmune) response results in graft rejection, which is manifested as deterioration or complete loss of graft function. In contrast, autoimmunity is an immune response to the self's own antigens. Alloimmunization (isoimmunization) is the process of becoming alloimmune, that is, developing the relevant antibodies for the first time.

A serotype or serovar is a distinct variation within a species of bacteria or virus or among immune cells of different individuals. These microorganisms, viruses, or cells are classified together based on their surface antigens, allowing the epidemiologic classification of organisms to the subspecies level. A group of serovars with common antigens is called a serogroup or sometimes serocomplex.

Jan Klein | Czech - American immunologist | January 18, 1936 - May 7, 2023

HLA-DR is an MHC class II cell surface receptor encoded by the human leukocyte antigen complex on chromosome 6 region 6p21.31. The complex of HLA-DR and peptide, generally between 9 and 30 amino acids in length, constitutes a ligand for the T-cell receptor (TCR). HLA were originally defined as cell surface antigens that mediate graft-versus-host disease. Identification of these antigens has led to greater success and longevity in organ transplant.

MHC Class II molecules are a class of major histocompatibility complex (MHC) molecules normally found only on professional antigen-presenting cells such as dendritic cells, mononuclear phagocytes, some endothelial cells, thymic epithelial cells, and B cells. These cells are important in initiating immune responses.

HLA-DQ (DQ) is a cell surface receptor protein found on antigen-presenting cells. It is an αβ heterodimer of type MHC class II. The α and β chains are encoded by two loci, HLA-DQA1 and HLA-DQB1, that are adjacent to each other on chromosome band 6p21.3. Both α-chain and β-chain vary greatly. A person often produces two α-chain and two β-chain variants and thus 4 isoforms of DQ. The DQ loci are in close genetic linkage to HLA-DR, and less closely linked to HLA-DP, HLA-A, HLA-B and HLA-C.
HLA-DP is a protein/peptide-antigen receptor and graft-versus-host disease antigen that is composed of 2 subunits, DPα and DPβ. DPα and DPβ are encoded by two loci, HLA-DPA1 and HLA-DPB1, that are found in the MHC Class II region in the Human Leukocyte Antigen complex on human chromosome 6 . Less is known about HLA-DP relative to HLA-DQ and HLA-DR but the sequencing of DP types and determination of more frequent haplotypes has progressed greatly within the last few years.
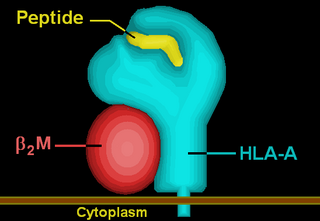
HLA-A is a group of human leukocyte antigens (HLA) that are encoded by the HLA-A locus, which is located at human chromosome 6p21.3. HLA is a major histocompatibility complex (MHC) antigen specific to humans. HLA-A is one of three major types of human MHC class I transmembrane proteins. The others are HLA-B and HLA-C. The protein is a heterodimer, and is composed of a heavy α chain and smaller β chain. The α chain is encoded by a variant HLA-A gene, and the β chain (β2-microglobulin) is an invariant β2 microglobulin molecule. The β2 microglobulin protein is encoded by the B2M gene, which is located at chromosome 15q21.1 in humans.
Tissue typing is a procedure in which the tissues of a prospective donor and recipient are tested for compatibility prior to transplantation. Mismatched donor and recipient tissues can lead to rejection of the tissues. There are multiple methods of tissue typing.

Minor histocompatibility antigen are peptides presented on the cellular surface of donated organs that are known to give an immunological response in some organ transplants. They cause problems of rejection less frequently than those of the major histocompatibility complex (MHC). Minor histocompatibility antigens (MiHAs) are diverse, short segments of proteins and are referred to as peptides. These peptides are normally around 9-12 amino acids in length and are bound to both the major histocompatibility complex (MHC) class I and class II proteins. Peptide sequences can differ among individuals and these differences arise from SNPs in the coding region of genes, gene deletions, frameshift mutations, or insertions. About a third of the characterized MiHAs come from the Y chromosome. Prior to becoming a short peptide sequence, the proteins expressed by these polymorphic or diverse genes need to be digested in the proteasome into shorter peptides. These endogenous or self peptides are then transported into the endoplasmic reticulum with a peptide transporter pump called TAP where they encounter and bind to the MHC class I molecule. This contrasts with MHC class II molecules's antigens which are peptides derived from phagocytosis/endocytosis and molecular degradation of non-self entities' proteins, usually by antigen-presenting cells. MiHA antigens are either ubiquitously expressed in most tissue like skin and intestines or restrictively expressed in the immune cells.

HLA-A*02 (A*02) is a human leukocyte antigen serotype within the HLA-A serotype group. The serotype is determined by the antibody recognition of the α2 domain of the HLA-A α-chain. For A*02, the α chain is encoded by the HLA-A*02 gene and the β chain is encoded by the B2M locus. In 2010 the World Health Organization Naming Committee for Factors of the HLA System revised the nomenclature for HLAs. Before this revision, HLA-A*02 was also referred to as HLA-A2, HLA-A02, and HLA-A*2.

HLA-G histocompatibility antigen, class I, G, also known as human leukocyte antigen G (HLA-G), is a protein that in humans is encoded by the HLA-G gene.
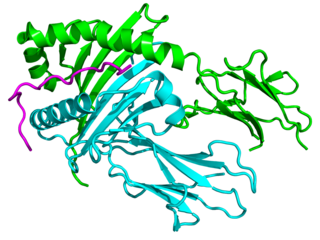
Major histocompatibility complex, class II, DQ alpha 1, also known as HLA-DQA1, is a human gene present on short arm of chromosome 6 (6p21.3) and also denotes the genetic locus which contains this gene. The protein encoded by this gene is one of two proteins that are required to form the DQ heterodimer, a cell surface receptor essential to the function of the immune system.
The following outline is provided as an overview of and topical guide to immunology:
Human leukocyte antigens (HLA) began as a list of antigens identified as a result of transplant rejection. The antigens were initially identified by categorizing and performing massive statistical analyses on interactions between blood types. This process is based upon the principle of serotypes. HLA are not typical antigens, like those found on surface of infectious agents. HLAs are alloantigens, they vary from individual to individual as a result of genetic differences. An organ called the thymus is responsible for ensuring that any T-cells that attack self proteins are not allowed to live. In essence, every individual's immune system is tuned to the specific set of HLA and self proteins produced by that individual; where this goes awry is when tissues are transferred to another person. Since individuals almost always have different "banks" of HLAs, the immune system of the recipient recognizes the transplanted tissue as non-self and destroys the foreign tissue, leading to transplant rejection. It was through the realization of this that HLAs were discovered.
Alloantigen recognition refers to immune system recognition of genetically encoded polymorphisms among the genetically distinguishable members of same species. Post-transplant recognition of alloantigens occurs in secondary lymphoid organs. Donor specific antigens are recognized by recipient’s T lymphocytes and triggers adaptive pro-inflammatory response which consequently leads to rejection of allogenic transplants. Allospecific T lymphocytes may be stimulated by three major pathways: direct recognition, indirect recognition or semidirect recognition. The pathway involved in specific cases is dictated by intrinsic and extrinsic factors of allograft and directly influence nature and magnitude of T lymphocytes mediated immune response. Furthermore, variant tissues and organs such as skin or cornea or solid organ transplants can be recognized in different pathways and therefore are rejected in different fashion.












