The major histocompatibility complex (MHC) is a large locus on vertebrate DNA containing a set of closely linked polymorphic genes that code for cell surface proteins essential for the adaptive immune system. These cell surface proteins are called MHC molecules.
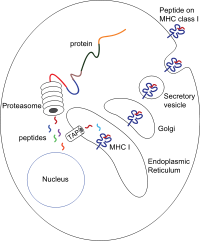
Antigen processing, or the cytosolic pathway, is an immunological process that prepares antigens for presentation to special cells of the immune system called T lymphocytes. It is considered to be a stage of antigen presentation pathways. This process involves two distinct pathways for processing of antigens from an organism's own (self) proteins or intracellular pathogens, or from phagocytosed pathogens ; subsequent presentation of these antigens on class I or class II major histocompatibility complex (MHC) molecules is dependent on which pathway is used. Both MHC class I and II are required to bind antigens before they are stably expressed on a cell surface. MHC I antigen presentation typically involves the endogenous pathway of antigen processing, and MHC II antigen presentation involves the exogenous pathway of antigen processing. Cross-presentation involves parts of the exogenous and the endogenous pathways but ultimately involves the latter portion of the endogenous pathway.

Bare lymphocyte syndrome is a condition caused by mutations in certain genes of the major histocompatibility complex or involved with the processing and presentation of MHC molecules. It is a form of severe combined immunodeficiency.

Antigen presentation is a vital immune process that is essential for T cell immune response triggering. Because T cells recognize only fragmented antigens displayed on cell surfaces, antigen processing must occur before the antigen fragment can be recognized by a T-cell receptor. Specifically, the fragment, bound to the major histocompatibility complex (MHC), is transported to the surface of the cell, a process known as presentation. If there has been an infection with viruses or bacteria, the cell will present an endogenous or exogenous peptide fragment derived from the antigen by MHC molecules. There are two types of MHC molecules which differ in the behaviour of the antigens: MHC class I molecules (MHC-I) bind peptides from the cell cytosol, while peptides generated in the endocytic vesicles after internalisation are bound to MHC class II (MHC-II). Cellular membranes separate these two cellular environments - intracellular and extracellular. Each T cell can only recognize tens to hundreds of copies of a unique sequence of a single peptide among thousands of other peptides presented on the same cell, because an MHC molecule in one cell can bind to quite a large range of peptides. Predicting which antigens will be presented to the immune system by a certain MHC/HLA type is difficult, but the technology involved is improving.

MHC Class II molecules are a class of major histocompatibility complex (MHC) molecules normally found only on professional antigen-presenting cells such as dendritic cells, mononuclear phagocytes, some endothelial cells, thymic epithelial cells, and B cells. These cells are important in initiating immune responses.
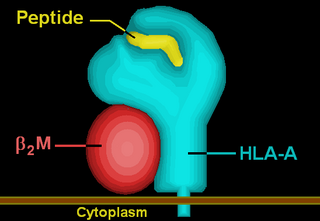
HLA-A is a group of human leukocyte antigens (HLA) that are encoded by the HLA-A locus, which is located at human chromosome 6p21.3. HLA is a major histocompatibility complex (MHC) antigen specific to humans. HLA-A is one of three major types of human MHC class I transmembrane proteins. The others are HLA-B and HLA-C. The protein is a heterodimer, and is composed of a heavy α chain and smaller β chain. The α chain is encoded by a variant HLA-A gene, and the β chain (β2-microglobulin) is an invariant β2 microglobulin molecule. The β2 microglobulin protein is encoded by the B2M gene, which is located at chromosome 15q21.1 in humans.

CLIP or Class II-associated invariant chain peptide is the part of the invariant chain (Ii) that binds to the peptide binding groove of MHC class II and remains there until the MHC receptor is fully assembled. CLIP is one of the most prevalent self peptides found in the thymic cortex of most antigen-presenting cells. The purpose of CLIP is to prevent the degradation of MHC II dimers before antigenic peptides bind, and to prevent autoimmunity.

HLA class I histocompatibility antigen, alpha chain E (HLA-E) also known as MHC class I antigen E is a protein that in humans is encoded by the HLA-E gene. The human HLA-E is a non-classical MHC class I molecule that is characterized by a limited polymorphism and a lower cell surface expression than its classical paralogues. The functional homolog in mice is called Qa-1b, officially known as H2-T23.

Minor histocompatibility antigen are peptides presented on the cellular surface of donated organs that are known to give an immunological response in some organ transplants. They cause problems of rejection less frequently than those of the major histocompatibility complex (MHC). Minor histocompatibility antigens (MiHAs) are diverse, short segments of proteins and are referred to as peptides. These peptides are normally around 9-12 amino acids in length and are bound to both the major histocompatibility complex (MHC) class I and class II proteins. Peptide sequences can differ among individuals and these differences arise from SNPs in the coding region of genes, gene deletions, frameshift mutations, or insertions. About a third of the characterized MiHAs come from the Y chromosome. Prior to becoming a short peptide sequence, the proteins expressed by these polymorphic or diverse genes need to be digested in the proteasome into shorter peptides. These endogenous or self peptides are then transported into the endoplasmic reticulum with a peptide transporter pump called TAP where they encounter and bind to the MHC class I molecule. This contrasts with MHC class II molecules's antigens which are peptides derived from phagocytosis/endocytosis and molecular degradation of non-self entities' proteins, usually by antigen-presenting cells. MiHA antigens are either ubiquitously expressed in most tissue like skin and intestines or restrictively expressed in the immune cells.

TAP-associated glycoprotein, also known as tapasin or TAPBP, is a protein that in humans is encoded by the TAPBP gene.

Protein disulfide-isomerase A3 (PDIA3), also known as glucose-regulated protein, 58-kD (GRP58), is an isomerase enzyme encoded by the autosomal gene PDIA3 in humans. This protein localizes to the endoplasmic reticulum (ER) and interacts with lectin chaperones calreticulin and calnexin (CNX) to modulate folding of newly synthesized glycoproteins. It is thought that complexes of lectins and this protein mediate protein folding by promoting formation of disulfide bonds in their glycoprotein substrates.

HLA-DM is an intracellular protein involved in the mechanism of antigen presentation on antigen presenting cells (APCs) of the immune system. It does this by assisting in peptide loading of major histocompatibility complex (MHC) class II membrane-bound proteins. HLA-DM is encoded by the genes HLA-DMA and HLA-DMB.

Transporter associated with antigen processing 1 (TAP1) is a protein that in humans is encoded by the TAP1 gene. A member of the ATP-binding cassette transporter family, it is also known as ABCB2.
HLA class II histocompatibility antigen gamma chain also known as HLA-DR antigens-associated invariant chain or CD74, is a protein that in humans is encoded by the CD74 gene. The invariant chain is a polypeptide which plays a critical role in antigen presentation. It is involved in the formation and transport of MHC class II peptide complexes for the generation of CD4+ T cell responses. The cell surface form of the invariant chain is known as CD74. CD74 is a cell surface receptor for the cytokine macrophage migration inhibitory factor (MIF).

HLA class I histocompatibility antigen, alpha chain F is a protein that in humans is encoded by the HLA-F gene. It is an empty intracellular molecule that encodes a non-classical heavy chain anchored to the membrane and forming a heterodimer with a β-2 microglobulin light chain. It belongs to the HLA class I heavy chain paralogues that separate from most of the HLA heavy chains. HLA-F is localized in the endoplasmic reticulum and Golgi apparatus, and is also unique in the sense that it exhibits few polymorphisms in the human population relative to the other HLA genes; however, there have been found different isoforms from numerous transcript variants found for the HLA-F gene. Its pathways include IFN-gamma signaling and CDK-mediated phosphorylation and removal of the Saccharomycescerevisiae Cdc6 protein, which is crucial for functional DNA replication.
Type 1 tumor necrosis factor receptor shedding aminopeptidase regulator, also known as endoplasmic reticulum aminopeptidase 1 (ARTS-1), is a protein which in humans is encoded by the ARTS-1 gene.

Minor histocompatibility antigen H13 is a protein that in humans is encoded by the HM13 gene.

TAP2 is a gene in humans that encodes the protein Antigen peptide transporter 2.
Immunoevasins are proteins expressed by some viruses that enable the virus to evade immune recognition by interfering with MHC I complexes in the infected cell, therefore blocking the recognition of viral protein fragments by CD8+ cytotoxic T lymphocytes. Less frequently, MHC II antigen presentation and induced-self molecules may also be targeted. Some viral immunoevasins block peptide entry into the endoplasmic reticulum (ER) by targeting the TAP transporters. Immunoevasins are particularly abundant in viruses that are capable of establishing long-term infections of the host, such as herpesviruses.

The peptide-loading complex (PLC) is a short-lived, multisubunit membrane protein complex that is located in the endoplasmic reticulum (ER). It orchestrates peptide translocation and selection by major histocompatibility complex class I (MHC-I) molecules. Stable peptide-MHC I complexes are released to the cell surface to promote T-cell response against malignant or infected cells. In turn, T-cells recognize the activated peptides, which could be immunogenic or non-immunogenic.