
In genetics and bioinformatics, a single-nucleotide polymorphism is a germline substitution of a single nucleotide at a specific position in the genome that is present in a sufficiently large fraction of considered population.
The International HapMap Project was an organization that aimed to develop a haplotype map (HapMap) of the human genome, to describe the common patterns of human genetic variation. HapMap is used to find genetic variants affecting health, disease and responses to drugs and environmental factors. The information produced by the project is made freely available for research.
Researchers have investigated the relationship between race and genetics as part of efforts to understand how biology may or may not contribute to human racial categorization. Today, the consensus among scientists is that race is a social construct, and that using it as a proxy for genetic differences among populations is misleading.

A DNA segment is identical by state (IBS) in two or more individuals if they have identical nucleotide sequences in this segment. An IBS segment is identical by descent (IBD) in two or more individuals if they have inherited it from a common ancestor without recombination, that is, the segment has the same ancestral origin in these individuals. DNA segments that are IBD are IBS per definition, but segments that are not IBD can still be IBS due to the same mutations in different individuals or recombinations that do not alter the segment.
Genetics and archaeogenetics of South Asia is the study of the genetics and archaeogenetics of the ethnic groups of South Asia. It aims at uncovering these groups' genetic histories. The geographic position of the Indian subcontinent makes its biodiversity important for the study of the early dispersal of anatomically modern humans across Asia.

In population genetics, an ancestry-informative marker (AIM) is a single-nucleotide polymorphism that exhibits substantially different frequencies between different populations. A set of many AIMs can be used to estimate the proportion of ancestry of an individual derived from each population.
Neil Risch is an American human geneticist and professor at the University of California, San Francisco (UCSF). Risch is the Lamond Family Foundation Distinguished Professor in Human Genetics, Founding Director of the Institute for Human Genetics, and Professor of Epidemiology and Biostatistics at UCSF. He specializes in statistical genetics, genetic epidemiology and population genetics.

Human genetic variation is the genetic differences in and among populations. There may be multiple variants of any given gene in the human population (alleles), a situation called polymorphism.
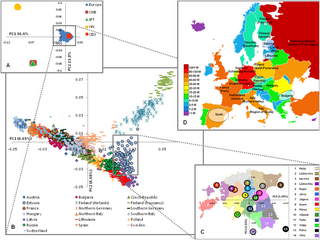
The genetic history of Europe includes information around the formation, ethnogenesis, and other DNA-specific information about populations indigenous, or living in Europe.
Population genomics is the large-scale comparison of DNA sequences of populations. Population genomics is a neologism that is associated with population genetics. Population genomics studies genome-wide effects to improve our understanding of microevolution so that we may learn the phylogenetic history and demography of a population.

The genetic history of the Middle East is the subject of research within the fields of human population genomics, archaeogenetics and Middle Eastern studies. Researchers use Y-DNA, mtDNA, and other autosomal DNAs to identify the genetic history of ancient and modern populations of Egypt, Persia, Mesopotamia, Anatolia, Arabia, the Levant, and other areas.
Population structure is the presence of a systematic difference in allele frequencies between subpopulations. In a randomly mating population, allele frequencies are expected to be roughly similar between groups. However, mating tends to be non-random to some degree, causing structure to arise. For example, a barrier like a river can separate two groups of the same species and make it difficult for potential mates to cross; if a mutation occurs, over many generations it can spread and become common in one subpopulation while being completely absent in the other.
Genetic studies of Jews are part of the population genetics discipline and are used to analyze the chronology of Jewish migration accompanied by research in other fields, such as history, linguistics, archaeology, and paleontology. These studies investigate the origins of various Jewish ethnic divisions. In particular, they examine whether there is a common genetic heritage among them. The medical genetics of Jews are studied for population-specific diseases.
Population genetics research has been conducted on the Turkish people, who comprise the demographic majority of Turkey. Such studies are relevant for the demographic history of the population as well as health reasons, such as population-specific diseases. Some studies have sought to determine the relative contributions of the Turkic peoples of Central Asia, from where the Seljuk Turks began migrating to Anatolia after the Battle of Manzikert in 1071, which led to the establishment of the Anatolian Seljuk Sultanate in the late 11th century, and prior populations in the area who were Turkified during the Seljuk and Ottoman periods.
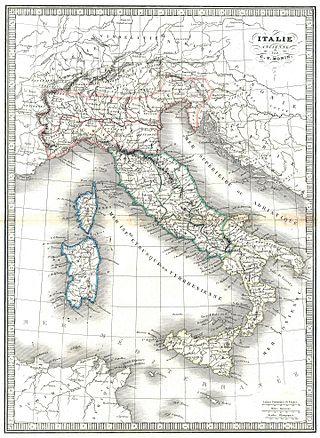
The genetic history of Italy is greatly influenced by geography and history. The ancestors of Italians were mostly Indo-European speaking peoples and pre-Indo-European speakers. During the Roman empire, the city of Rome also attracted people from various regions of the Mediterranean basin, including Southern Europe, North Africa and the Middle East. Based on DNA analysis, there is evidence of ancient regional genetic substructure and continuity within modern Italy dating to the pre-Roman and Roman periods.
Jonathan Karl Pritchard is an English-born professor of genetics at Stanford University, best known for his development of the STRUCTURE algorithm for studying population structure and his work on human genetic variation and evolution. His research interests lie in the study of human evolution, in particular in understanding the association between genetic variation among human individuals and human traits.
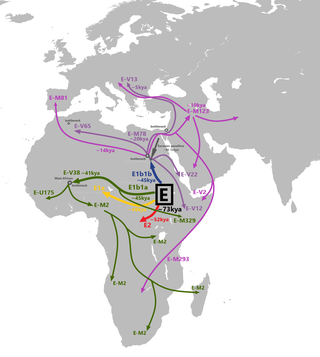
The genetic history of Egypt reflects its geographical location at the crossroads of several major biocultural areas: North Africa, the Sahara, the Middle East, the Mediterranean and sub-Saharan Africa.
The study of the genetics and archaeogenetics of the Gujarati people of India aims at uncovering these people's genetic history. According to the 1000 Genomes Project, "Gujarati" is a general term used to describe people who trace their ancestry to the region of Gujarat, located in the northwestern part of the Indian subcontinent, and who speak the Gujarati language, an Indo-European language. They have some genetic commonalities as well as differences with other ethnic groups of India.

In archaeogenetics, the term Western Hunter-Gatherer (WHG), West European Hunter-Gatherer, Western European Hunter-Gatherer, Villabruna cluster, or Oberkassel cluster is the name given to a distinct ancestral component of modern Europeans, representing descent from a population of Mesolithic hunter-gatherers who scattered over Western, Southern and Central Europe, from the British Isles in the west to the Carpathians in the east, following the retreat of the ice sheet of the Last Glacial Maximum.

The genetic history of Africa is composed of the overall genetic history of African populations in Africa, including the regional genetic histories of North Africa, West Africa, East Africa, Central Africa, and Southern Africa, as well as the recent origin of modern humans in Africa. The Sahara served as a trans-regional passageway and place of dwelling for people in Africa during various humid phases and periods throughout the history of Africa.











