
Histone acetyltransferases (HATs) are enzymes that acetylate conserved lysine amino acids on histone proteins by transferring an acetyl group from acetyl-CoA to form ε-N-acetyllysine. DNA is wrapped around histones, and, by transferring an acetyl group to the histones, genes can be turned on and off. In general, histone acetylation increases gene expression.

Histone deacetylases (EC 3.5.1.98, HDAC) are a class of enzymes that remove acetyl groups (O=C-CH3) from an ε-N-acetyl lysine amino acid on both histone and non-histone proteins. HDACs allow histones to wrap the DNA more tightly. This is important because DNA is wrapped around histones, and DNA expression is regulated by acetylation and de-acetylation. HDAC's action is opposite to that of histone acetyltransferase. HDAC proteins are now also called lysine deacetylases (KDAC), to describe their function rather than their target, which also includes non-histone proteins. In general, they suppress gene expression.

Histone acetyltransferase p300 also known as p300 HAT or E1A-associated protein p300 also known as EP300 or p300 is an enzyme that, in humans, is encoded by the EP300 gene. It functions as histone acetyltransferase that regulates transcription of genes via chromatin remodeling by allowing histone proteins to wrap DNA less tightly. This enzyme plays an essential role in regulating cell growth and division, prompting cells to mature and assume specialized functions (differentiate), and preventing the growth of cancerous tumors. The p300 protein appears to be critical for normal development before and after birth.

Transcription factor Sp1, also known as specificity protein 1* is a protein that in humans is encoded by the SP1 gene.
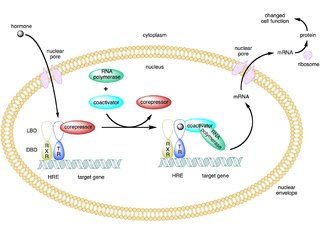
A coactivator is a type of transcriptional coregulator that binds to an activator to increase the rate of transcription of a gene or set of genes. The activator contains a DNA binding domain that binds either to a DNA promoter site or a specific DNA regulatory sequence called an enhancer. Binding of the activator-coactivator complex increases the speed of transcription by recruiting general transcription machinery to the promoter, therefore increasing gene expression. The use of activators and coactivators allows for highly specific expression of certain genes depending on cell type and developmental stage.

P300/CBP-associated factor (PCAF), also known as K(lysine) acetyltransferase 2B (KAT2B), is a human gene and transcriptional coactivator associated with p53.

CREB-binding protein, also known as CREBBP or CBP or KAT3A, is a coactivator encoded by the CREBBP gene in humans, located on chromosome 16p13.3. CBP has intrinsic acetyltransferase functions; it is able to add acetyl groups to both transcription factors as well as histone lysines, the latter of which has been shown to alter chromatin structure making genes more accessible for transcription. This relatively unique acetyltransferase activity is also seen in another transcription enzyme, EP300 (p300). Together, they are known as the p300-CBP coactivator family and are known to associate with more than 16,000 genes in humans; however, while these proteins share many structural features, emerging evidence suggests that these two co-activators may promote transcription of genes with different biological functions.

Histone acetylation and deacetylation are the processes by which the lysine residues within the N-terminal tail protruding from the histone core of the nucleosome are acetylated and deacetylated as part of gene regulation.
Chromatin remodeling is the dynamic modification of chromatin architecture to allow access of condensed genomic DNA to the regulatory transcription machinery proteins, and thereby control gene expression. Such remodeling is principally carried out by 1) covalent histone modifications by specific enzymes, e.g., histone acetyltransferases (HATs), deacetylases, methyltransferases, and kinases, and 2) ATP-dependent chromatin remodeling complexes which either move, eject or restructure nucleosomes. Besides actively regulating gene expression, dynamic remodeling of chromatin imparts an epigenetic regulatory role in several key biological processes, egg cells DNA replication and repair; apoptosis; chromosome segregation as well as development and pluripotency. Aberrations in chromatin remodeling proteins are found to be associated with human diseases, including cancer. Targeting chromatin remodeling pathways is currently evolving as a major therapeutic strategy in the treatment of several cancers.

Transformation/transcription domain-associated protein, also known as TRRAP, is a protein that in humans is encoded by the TRRAP gene. TRRAP belongs to the phosphatidylinositol 3-kinase-related kinase protein family.

Histone acetyltransferase KAT2A is an enzyme that in humans is encoded by the KAT2A gene.

Transcription initiation factor TFIID subunit 10 is a protein that in humans is encoded by the TAF10 gene.

Histone H4 is a protein that in humans is encoded by the HIST2H4A gene.

Histone H2A type 2-C is a protein that in humans is encoded by the HIST2H2AC gene.

Histone acetyltransferase KAT7 is an enzyme that in humans is encoded by the KAT7 gene. It specifically acetylates H4 histones at the lysine12 residue (H4K12) and is necessary for origin licensing and DNA replication. KAT7 associates with origins of replication during G1 phase of the cell cycle through complexing with CDT1. Geminin is thought to inhibit the acetyltransferase activity of KAT7 when KAT7 and CDT1 are complexed together.
Histone H3.1 is a protein that in humans is encoded by the HIST1H3C gene.

K(lysine) acetyltransferase 8 (KAT8) is an enzyme that in humans is encoded by the KAT8 gene.

JADE1 is a protein that in humans is encoded by the JADE1 gene.
While the cellular and molecular mechanisms of learning and memory have long been a central focus of neuroscience, it is only in recent years that attention has turned to the epigenetic mechanisms behind the dynamic changes in gene transcription responsible for memory formation and maintenance. Epigenetic gene regulation often involves the physical marking of DNA or associated proteins to cause or allow long-lasting changes in gene activity. Epigenetic mechanisms such as DNA methylation and histone modifications have been shown to play an important role in learning and memory.
The NuA4 histone acetyltransferase complex is a protein complex that has histone acetylase activity on chromatin, as well as ATPase, DNA helicase and structural DNA binding activities. The complex is thought to be involved in double-strand DNA break repair. Subunits of the human complex include HTATIP/TIP60, TRRAP, RUVBL1, RUVBL2, beta-actin and BAF53/ACTL6A. In yeast, the complex has 13 subunits, including the catalytic subunit Esa1.

























