
An antibody (Ab), also known as an immunoglobulin (Ig), is a large, Y-shaped protein used by the immune system to identify and neutralize foreign objects such as pathogenic bacteria and viruses. The antibody recognizes a unique molecule of the pathogen, called an antigen. Each tip of the "Y" of an antibody contains a paratope that is specific for one particular epitope on an antigen, allowing these two structures to bind together with precision. Using this binding mechanism, an antibody can tag a microbe or an infected cell for attack by other parts of the immune system, or can neutralize it directly.
Glycomics is the comprehensive study of glycomes, including genetic, physiologic, pathologic, and other aspects. Glycomics "is the systematic study of all glycan structures of a given cell type or organism" and is a subset of glycobiology. The term glycomics is derived from the chemical prefix for sweetness or a sugar, "glyco-", and was formed to follow the omics naming convention established by genomics and proteomics.

A polyhistidine-tag is an amino acid motif in proteins that typically consists of at least six histidine (His) residues, often at the N- or C-terminus of the protein. It is also known as hexa histidine-tag, 6xHis-tag, His6 tag, by the US trademarked name HIS TAG, and most commonly as His-Tag. The tag was invented by Roche, although the use of histidines and its vectors are distributed by Qiagen. Various purification kits for histidine-tagged proteins are available from Qiagen, Sigma, Thermo Scientific, GE Healthcare, Macherey-Nagel, Cube Biotech, Clontech, Bio-Rad, and others.
Protein purification is a series of processes intended to isolate one or a few proteins from a complex mixture, usually cells, tissues or whole organisms. Protein purification is vital for the specification of the function, structure and interactions of the protein of interest. The purification process may separate the protein and non-protein parts of the mixture, and finally separate the desired protein from all other proteins. Ideally, to study a protein of interest, it must be separated from other components of the cell so that contaminants won't interfere in the examination of the protein of interest's structure and function. Separation of one protein from all others is typically the most laborious aspect of protein purification. Separation steps usually exploit differences in protein size, physico-chemical properties, binding affinity and biological activity. The pure result may be termed protein isolate.
In biochemistry, biotinylation is the process of covalently attaching biotin to a protein, nucleic acid or other molecule. Biotinylation is rapid, specific and is unlikely to disturb the natural function of the molecule due to the small size of biotin. Biotin binds to streptavidin and avidin with an extremely high affinity, fast on-rate, and high specificity, and these interactions are exploited in many areas of biotechnology to isolate biotinylated molecules of interest. Biotin-binding to streptavidin and avidin is resistant to extremes of heat, pH and proteolysis, making capture of biotinylated molecules possible in a wide variety of environments. Also, multiple biotin molecules can be conjugated to a protein of interest, which allows binding of multiple streptavidin, avidin or neutravidin protein molecules and increases the sensitivity of detection of the protein of interest. There is a large number of biotinylation reagents available that exploit the wide range of possible labelling methods. Due to the strong affinity between biotin and streptavidin, the purification of biotinylated proteins has been a widely used approach to identify protein-protein interactions and post-translational events such as ubiquitylation in molecular biology.

Phage display is a laboratory technique for the study of protein–protein, protein–peptide, and protein–DNA interactions that uses bacteriophages to connect proteins with the genetic information that encodes them. In this technique, a gene encoding a protein of interest is inserted into a phage coat protein gene, causing the phage to "display" the protein on its outside while containing the gene for the protein on its inside, resulting in a connection between genotype and phenotype. These displaying phages can then be screened against other proteins, peptides or DNA sequences, in order to detect interaction between the displayed protein and those other molecules. In this way, large libraries of proteins can be screened and amplified in a process called in vitro selection, which is analogous to natural selection.
Affinity chromatography is a method of separating a biomolecule from a mixture, based on a highly specific macromolecular binding interaction between the biomolecule and another substance. The specific type of binding interaction depends on the biomolecule of interest; antigen and antibody, enzyme and substrate, receptor and ligand, or protein and nucleic acid binding interactions are frequently exploited for isolation of various biomolecules. Affinity chromatography is useful for its high selectivity and resolution of separation, compared to other chromatographic methods.
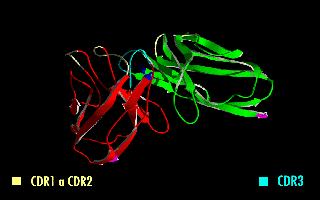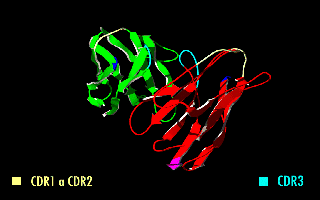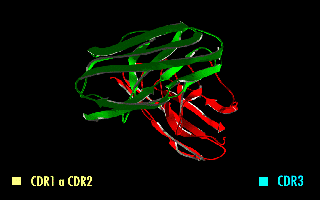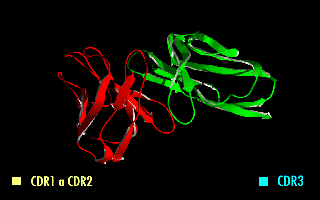
A single-chain variable fragment (scFv) is not actually a fragment of an antibody, but instead is a fusion protein of the variable regions of the heavy (VH) and light chains (VL) of immunoglobulins, connected with a short linker peptide of ten to about 25 amino acids. The linker is usually rich in glycine for flexibility, as well as serine or threonine for solubility, and can either connect the N-terminus of the VH with the C-terminus of the VL, or vice versa. This protein retains the specificity of the original immunoglobulin, despite removal of the constant regions and the introduction of the linker. The image to the right shows how this modification usually leaves the specificity unaltered.
A protein microarray is a high-throughput method used to track the interactions and activities of proteins, and to determine their function, and determining function on a large scale. Its main advantage lies in the fact that large numbers of proteins can be tracked in parallel. The chip consists of a support surface such as a glass slide, nitrocellulose membrane, bead, or microtitre plate, to which an array of capture proteins is bound. Probe molecules, typically labeled with a fluorescent dye, are added to the array. Any reaction between the probe and the immobilised protein emits a fluorescent signal that is read by a laser scanner. Protein microarrays are rapid, automated, economical, and highly sensitive, consuming small quantities of samples and reagents. The concept and methodology of protein microarrays was first introduced and illustrated in antibody microarrays in 1983 in a scientific publication and a series of patents. The high-throughput technology behind the protein microarray was relatively easy to develop since it is based on the technology developed for DNA microarrays, which have become the most widely used microarrays.
Protein tags are peptide sequences genetically grafted onto a recombinant protein. Tags are attached to proteins for various purposes. They can be added to either end of the target protein, so they are either C-terminus or N-terminus specific or are both C-terminus and N-terminus specific. Some tags are also inserted at sites within the protein of interest; they are known as internal tags.
Bacterial display is a protein engineering technique used for in vitro protein evolution. Libraries of polypeptides displayed on the surface of bacteria can be screened using flow cytometry or iterative selection procedures (biopanning). This protein engineering technique allows us to link the function of a protein with the gene that encodes it. Bacterial display can be used to find target proteins with desired properties and can be used to make affinity ligands which are cell-specific. This system can be used in many applications including the creation of novel vaccines, the identification of enzyme substrates and finding the affinity of a ligand for its target protein.

Protein A is a 42 kDa surface protein originally found in the cell wall of the bacteria Staphylococcus aureus. It is encoded by the spa gene and its regulation is controlled by DNA topology, cellular osmolarity, and a two-component system called ArlS-ArlR. It has found use in biochemical research because of its ability to bind immunoglobulins. It is composed of five homologous Ig-binding domains that fold into a three-helix bundle. Each domain is able to bind proteins from many mammalian species, most notably IgGs. It binds the heavy chain within the Fc region of most immunoglobulins and also within the Fab region in the case of the human VH3 family. Through these interactions in serum, where IgG molecules are bound in the wrong orientation, the bacteria disrupts opsonization and phagocytosis.
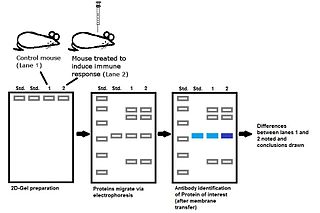
Immunoproteomics is the study of large sets of proteins (proteomics) involved in the immune response.

Meir Wilchek is an Israeli biochemist. He is a professor at the Weizmann Institute of Science.
Affibody molecules are small, robust proteins engineered to bind to a large number of target proteins or peptides with high affinity, imitating monoclonal antibodies, and are therefore a member of the family of antibody mimetics. Affibody molecules are used in biochemical research and are being developed as potential new biopharmaceutical drugs. These molecules can be used for molecular recognition in diagnostic and therapeutic applications.
Antibody mimetics are organic compounds that, like antibodies, can specifically bind antigens, but that are not structurally related to antibodies. They are usually artificial peptides or proteins with a molar mass of about 3 to 20 kDa.
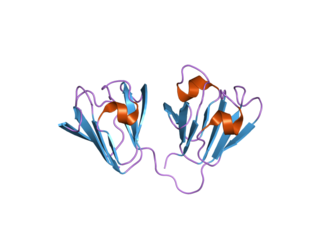
Affilins are artificial proteins designed to selectively bind antigens. Affilin proteins are structurally derived from human ubiquitin. Affilin proteins are constructed by modification of surface-exposed amino acids of these proteins and isolated by display techniques such as phage display and screening. They resemble antibodies in their affinity and specificity to antigens but not in structure, which makes them a type of antibody mimetic. Affilin was developed by Scil Proteins GmbH as potential new biopharmaceutical drugs, diagnostics and affinity ligands.
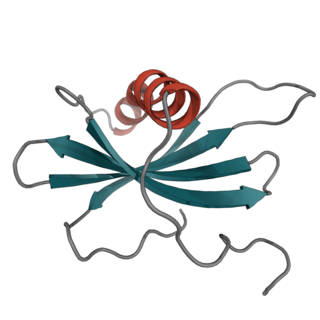
Affimer molecules are small proteins that bind to target proteins with affinity in the nanomolar range. These engineered non-antibody binding proteins are designed to mimic the molecular recognition characteristics of monoclonal antibodies in different applications. These affinity reagents have been optimized to increase their stability, make them tolerant to a range of temperatures and pH, reduce their size, and to increase their expression in E.coli and mammalian cells.
In the medical field of immunology, nanoCLAMP affinity reagents are recombinant 15 kD antibody mimetic proteins selected for tight, selective and gently reversible binding to target molecules. The nanoCLAMP scaffold is based on an IgG-like, thermostable carbohydrate binding module family 32 (CBM32) from a Clostridium perfringens hyaluronidase. The shape of nanoCLAMPs approximates a cylinder of approximately 4 nm in length and 2.5 nm in diameter, roughly the same size as a nanobody. nanoCLAMPs to specific targets are generated by varying the amino acid sequences and sometimes the length of three solvent exposed, adjacent loops that connect the beta strands making up the beta-sandwich fold, conferring binding affinity and specificity for the target.

IBA Lifesciences is a biotechnology company providing products and custom specific services for life science applications in academia and industry worldwide. IBA focusses on two business segments: cell selection and protein purification.











