
Oxidative phosphorylation or electron transport-linked phosphorylation or terminal oxidation is the metabolic pathway in which cells use enzymes to oxidize nutrients, thereby releasing chemical energy in order to produce adenosine triphosphate (ATP). In eukaryotes, this takes place inside mitochondria. Almost all aerobic organisms carry out oxidative phosphorylation. This pathway is so pervasive because it releases more energy than alternative fermentation processes such as anaerobic glycolysis.

An electron transport chain (ETC) is a series of protein complexes and other molecules that transfer electrons from electron donors to electron acceptors via redox reactions (both reduction and oxidation occurring simultaneously) and couples this electron transfer with the transfer of protons (H+ ions) across a membrane. The electrons that are transferred from NADH and FADH2 to the ETC involves four multi-subunit large enzymes complexes and two mobile electron carriers. Many of the enzymes in the electron transport chain are embedded within the membrane.

The cytochrome complex, or cyt c, is a small hemeprotein found loosely associated with the inner membrane of the mitochondrion where it plays a critical role in cellular respiration. It transfers electrons between Complexes III and IV. Cytochrome c is highly water-soluble, unlike other cytochromes. It is capable of undergoing oxidation and reduction as its iron atom converts between the ferrous and ferric forms, but does not bind oxygen. It also plays a major role in cell apoptosis. In humans, cytochrome c is encoded by the CYCS gene.

The enzyme cytochrome c oxidase or Complex IV, is a large transmembrane protein complex found in bacteria, archaea, and the mitochondria of eukaryotes.
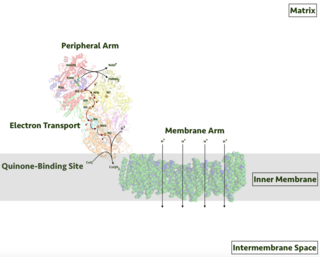
Respiratory complex I, EC 7.1.1.2 is the first large protein complex of the respiratory chains of many organisms from bacteria to humans. It catalyzes the transfer of electrons from NADH to coenzyme Q10 (CoQ10) and translocates protons across the inner mitochondrial membrane in eukaryotes or the plasma membrane of bacteria.

The coenzyme Q : cytochrome c – oxidoreductase, sometimes called the cytochrome bc1 complex, and at other times complex III, is the third complex in the electron transport chain, playing a critical role in biochemical generation of ATP. Complex III is a multisubunit transmembrane protein encoded by both the mitochondrial and the nuclear genomes. Complex III is present in the mitochondria of all animals and all aerobic eukaryotes and the inner membranes of most eubacteria. Mutations in Complex III cause exercise intolerance as well as multisystem disorders. The bc1 complex contains 11 subunits, 3 respiratory subunits, 2 core proteins and 6 low-molecular weight proteins.

In chemistry and biology, reactive oxygen species (ROS) are highly reactive chemicals formed from diatomic oxygen (O2), water, and hydrogen peroxide. Some prominent ROS are hydroperoxide (O2H), superoxide (O2-), hydroxyl radical (OH.), and singlet oxygen. ROS are pervasive because they are readily produced from O2, which is abundant. ROS are important in many ways, both beneficial and otherwise. ROS function as signals, that turn on and off biological functions. They are intermediates in the redox behavior of O2, which is central to fuel cells. ROS are central to the photodegradation of organic pollutants in the atmosphere. Most often however, ROS are discussed in a biological context, ranging from their effects on aging and their role in causing dangerous genetic mutations.
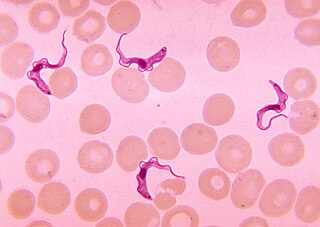
Trypanosoma is a genus of kinetoplastids, a monophyletic group of unicellular parasitic flagellate protozoa. Trypanosoma is part of the phylum Sarcomastigophora. The name is derived from the Greek trypano- (borer) and soma (body) because of their corkscrew-like motion. Most trypanosomes are heteroxenous and most are transmitted via a vector. The majority of species are transmitted by blood-feeding invertebrates, but there are different mechanisms among the varying species. Trypanosoma equiperdum is spread between horses and other equine species by sexual contact. They are generally found in the intestine of their invertebrate host, but normally occupy the bloodstream or an intracellular environment in the vertebrate host.

Trypanosoma brucei is a species of parasitic kinetoplastid belonging to the genus Trypanosoma that is present in sub-Saharan Africa. Unlike other protozoan parasites that normally infect blood and tissue cells, it is exclusively extracellular and inhabits the blood plasma and body fluids. It causes deadly vector-borne diseases: African trypanosomiasis or sleeping sickness in humans, and animal trypanosomiasis or nagana in cattle and horses. It is a species complex grouped into three subspecies: T. b. brucei, T. b. gambiense and T. b. rhodesiense. The first is a parasite of non-human mammals and causes nagana, while the latter two are zoonotic infecting both humans and animals and cause African trypanosomiasis.

The inner mitochondrial membrane (IMM) is the mitochondrial membrane which separates the mitochondrial matrix from the intermembrane space.
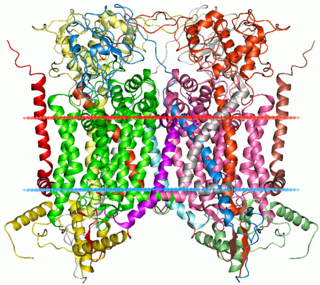
Cytochrome b within both molecular and cell biology, is a protein found in the mitochondria of eukaryotic cells. It functions as part of the electron transport chain and is the main subunit of transmembrane cytochrome bc1 and b6f complexes.
Thermogenic plants have the ability to raise their temperature above that of the surrounding air. Heat is generated in the mitochondria, as a secondary process of cellular respiration called thermogenesis. Alternative oxidase and uncoupling proteins similar to those found in mammals enable the process, which is still poorly understood.

Aldehyde oxidase (AO) is a metabolizing enzyme, located in the cytosolic compartment of tissues in many organisms. AO catalyzes the oxidation of aldehydes into carboxylic acid, and in addition, catalyzes the hydroxylation of some heterocycles. It can also catalyze the oxidation of both cytochrome P450 (CYP450) and monoamine oxidase (MAO) intermediate products. AO plays an important role in the metabolism of several drugs.

Ascofuranone is an antibiotic produced by various ascomycete fungi including Acremonium sclerotigenum that inhibits the Trypanosoma brucei alternative oxidase and is a lead compound in efforts to produce other drugs targeting this enzyme for the treatment of sleeping sickness. The compound is effective both in vitro cell culture and in infections in mice.

Cytochrome c oxidase subunit 5B, mitochondrial is an enzyme in humans that is a subunit of the cytochrome c oxidase complex, also known as Complex IV, the last enzyme in the mitochondrial electron transport chain. In humans, cytochrome c oxidase subunit 5B is encoded by the COX5B gene.

Salicylhydroxamic acid is a drug that is a potent and irreversible enzyme inhibitor of the urease enzyme in various bacteria and plants; it is usually used for urinary tract infections. The molecule is similar to urea but is not hydrolyzable by urease; it thus disrupts the bacteria's metabolism through competitive inhibition. It is also a trypanocidal agent. When administered orally, it is metabolized to salicylamide, which exerts analgesic, antipyretic, and anti-inflammatory effects.
The Arc system is a two-component system found in some bacteria that regulates gene expression in faculatative anaerobes such as Escheria coli. Two-component system means that it has a sensor molecule and a response regulator. Arc is an abbreviation for Anoxic Redox Control system. Arc systems are instrumental in maintaining energy metabolism during transcription of bacteria. The ArcA response regulator looks at growth conditions and expresses genes to best suit the bacteria. The Arc B sensor kinase, which is a tripartite protein, is membrane bound and can autophosphorylate.
Sulfide:quinone reductase is an enzyme with systematic name sulfide:quinone oxidoreductase. This enzyme catalyses the following chemical reaction
Plastid terminal oxidase or plastoquinol terminal oxidase (PTOX) is an enzyme that resides on the thylakoid membranes of plant and algae chloroplasts and on the membranes of cyanobacteria. The enzyme was hypothesized to exist as a photosynthetic oxidase in 1982 and was verified by sequence similarity to the mitochondrial alternative oxidase (AOX). The two oxidases evolved from a common ancestral protein in prokaryotes, and they are so functionally and structurally similar that a thylakoid-localized AOX can restore the function of a PTOX knockout.

Variant surface glycoprotein (VSG) is a ~60kDa protein which densely packs the cell surface of protozoan parasites belonging to the genus Trypanosoma. This genus is notable for their cell surface proteins. They were first isolated from Trypanosoma brucei in 1975 by George Cross. VSG allows the trypanosomatid parasites to evade the mammalian host's immune system by extensive antigenic variation. They form a 12–15 nm surface coat. VSG dimers make up ~90% of all cell surface protein and ~10% of total cell protein. For this reason, these proteins are highly immunogenic and an immune response raised against a specific VSG coat will rapidly kill trypanosomes expressing this variant. However, with each cell division there is a possibility that the progeny will switch expression to change the VSG that is being expressed. VSG has no prescribed biochemical activity.
















