
The β-sheet is a common motif of regular secondary structure in proteins. Beta sheets consist of beta strands connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet. A β-strand is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformation. The supramolecular association of β-sheets has been implicated in formation of the protein aggregates and fibrils observed in many human diseases, notably the amyloidoses such as Alzheimer's disease.

A transmembrane protein (TP) is a type of integral membrane protein that spans the entirety of the cell membrane. Many transmembrane proteins function as gateways to permit the transport of specific substances across the membrane. They frequently undergo significant conformational changes to move a substance through the membrane. They are usually highly hydrophobic and aggregate and precipitate in water. They require detergents or nonpolar solvents for extraction, although some of them (beta-barrels) can be also extracted using denaturing agents.
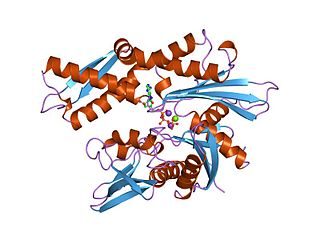
The 70 kilodalton heat shock proteins are a family of conserved ubiquitously expressed heat shock proteins. Proteins with similar structure exist in virtually all living organisms. The Hsp70s are an important part of the cell's machinery for protein folding, and help to protect cells from stress.
In biology and biochemistry, protease inhibitors, or antiproteases, are molecules that inhibit the function of proteases. Many naturally occurring protease inhibitors are proteins.

Hsp90 is a chaperone protein that assists other proteins to fold properly, stabilizes proteins against heat stress, and aids in protein degradation. It also stabilizes a number of proteins required for tumor growth, which is why Hsp90 inhibitors are investigated as anti-cancer drugs.
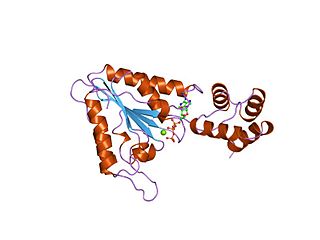
AAA proteins or ATPases Associated with diverse cellular Activities are a protein family sharing a common conserved module of approximately 230 amino acid residues. This is a large, functionally diverse protein family belonging to the AAA+ protein superfamily of ring-shaped P-loop NTPases, which exert their activity through the energy-dependent remodeling or translocation of macromolecules.
A supersecondary structure is a compact three-dimensional protein structure of several adjacent elements of a secondary structure that is smaller than a protein domain or a subunit. Supersecondary structures can act as nucleations in the process of protein folding.

Deubiquitinating enzymes (DUBs), also known as deubiquitinating peptidases, deubiquitinating isopeptidases, deubiquitinases, ubiquitin proteases, ubiquitin hydrolases, ubiquitin isopeptidases, are a large group of proteases that cleave ubiquitin from proteins. Ubiquitin is attached to proteins in order to regulate the degradation of proteins via the proteasome and lysosome; coordinate the cellular localisation of proteins; activate and inactivate proteins; and modulate protein-protein interactions. DUBs can reverse these effects by cleaving the peptide or isopeptide bond between ubiquitin and its substrate protein. In humans there are nearly 100 DUB genes, which can be classified into two main classes: cysteine proteases and metalloproteases. The cysteine proteases comprise ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), Machado-Josephin domain proteases (MJDs) and ovarian tumour proteases (OTU). The metalloprotease group contains only the Jab1/Mov34/Mpr1 Pad1 N-terminal+ (MPN+) (JAMM) domain proteases.

The TIM barrel is a conserved protein fold consisting of eight α-helices and eight parallel β-strands that alternate along the peptide backbone. The structure is named after triosephosphate isomerase, a conserved metabolic enzyme. TIM barrels are ubiquitous, with approximately 10% of all enzymes adopting this fold. Further, 5 of 7 enzyme commission (EC) enzyme classes include TIM barrel proteins. The TIM barrel fold is evolutionarily ancient, with many of its members possessing little similarity today, instead falling within the twilight zone of sequence similarity.

Pilin refers to a class of fibrous proteins that are found in pilus structures in bacteria. Bacterial pili are used in the exchange of genetic material during bacterial conjugation, while a shorter type of appendages also made up of pilin, called fimbriae, are used as a cell adhesion mechanism. Although not all bacteria have pili or fimbriae, bacterial pathogens often use their fimbriae to attach to host cells. In Gram-negative bacteria, where pili are more common, individual pilin molecules are linked by noncovalent protein-protein interactions, while Gram-positive bacteria often have polymerized pilin.

A leucine-rich repeat (LRR) is a protein structural motif that forms an α/β horseshoe fold. It is composed of repeating 20–30 amino acid stretches that are unusually rich in the hydrophobic amino acid leucine. These tandem repeats commonly fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Typically, each repeat unit has beta strand-turn-alpha helix structure, and the assembled domain, composed of many such repeats, has a horseshoe shape with an interior parallel beta sheet and an exterior array of helices. One face of the beta sheet and one side of the helix array are exposed to solvent and are therefore dominated by hydrophilic residues. The region between the helices and sheets is the protein's hydrophobic core and is tightly sterically packed with leucine residues.
The N-end rule is a rule that governs the rate of protein degradation through recognition of the N-terminal residue of proteins. The rule states that the N-terminal amino acid of a protein determines its half-life. The rule applies to both eukaryotic and prokaryotic organisms, but with different strength, rules, and outcome. In eukaryotic cells, these N-terminal residues are recognized and targeted by ubiquitin ligases, mediating ubiquitination thereby marking the protein for degradation. The rule was initially discovered by Alexander Varshavsky and co-workers in 1986. However, only rough estimations of protein half-life can be deduced from this 'rule', as N-terminal amino acid modification can lead to variability and anomalies, whilst amino acid impact can also change from organism to organism. Other degradation signals, known as degrons, can also be found in sequence.
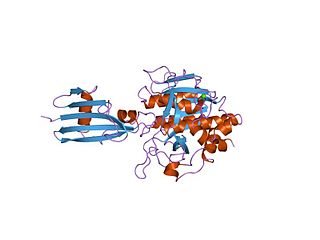
In molecular biology the protein SSI is a Subtilisin inhibitor-like which stands for Streptomyces subtilisin inhibitor. This is a protease inhibitor. These are often synthesised as part of a larger precursor protein, either as a prepropeptide. The function of this protein domain is to prevent access of the substrate to the active site. It is found only in bacteria.

In molecular biology, the AMMECR1 protein is a protein encoded by the AMMECR1 gene on human chromosome Xq22.3.
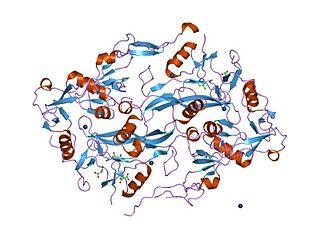
The Kazal domain is an evolutionary conserved protein domain usually indicative of serine protease inhibitors. However, kazal-like domains are also seen in the extracellular part of agrins, which are not known to be protease inhibitors.

In molecular biology, YopH, N-terminal refers to an evolutionary conserved protein domain. This entry represents the N-terminal domain of YopH protein tyrosine phosphatase (PTP).
Chaperone-usher fimbriae (CU) are linear, unbranching, outer-membrane pili secreted by gram-negative bacteria through the chaperone-usher system rather than through type IV secretion or extracellular nucleation systems. These fimbriae are built up out of modular pilus subunits, which are transported into the periplasm in a Sec dependent manner. Chaperone-usher secreted fimbriae are important pathogenicity factors facilitating host colonisation, localisation and biofilm formation in clinically important species such as uropathogenic Escherichia coli and Pseudomonas aeruginosa.

Acyldepsipeptide or cyclic acyldepsipeptide (ADEP) is a class of potential antibiotics first isolated from bacteria and act by deregulating the ClpP protease. Natural ADEPs were originally found as products of aerobic fermentation in Streptomyces hawaiiensis, A54556A and B, and in the culture broth of Streptomyces species, enopeptin A and B. ADEPs are of great interest in drug development due to their antibiotic properties and thus are being modified in attempt to achieve greater antimicrobial activity.

ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial is an enzyme that in humans is encoded by the CLPX gene. This protein is a member of the family of AAA Proteins and is to form the protein complex of Clp protease.

Cry6Aa is a toxic crystal protein generated by the bacterial family Bacillus thuringiensis during sporulation. This protein is a member of the alpha pore forming toxins family, which gives it insecticidal qualities advantageous in agricultural pest control. Each Cry protein has some level of target specificity; Cry6Aa has specific toxic action against coleopteran insects and nematodes. The corresponding B. thuringiensis gene, cry6aa, is located on bacterial plasmids. Along with several other Cry protein genes, cry6aa can be genetically recombined in Bt corn and Bt cotton so the plants produce specific toxins. Insects are developing resistance to the most commonly inserted proteins like Cry1Ac. Since Cry6Aa proteins function differently than other Cry proteins, they are combined with other proteins to decrease the development of pest resistance. Recent studies suggest this protein functions better in combination with other virulence factors such as other Cry proteins and metalloproteinases.
















