
Enterobacteria phage λ is a bacterial virus, or bacteriophage, that infects the bacterial species Escherichia coli. It was discovered by Esther Lederberg in 1950. The wild type of this virus has a temperate life cycle that allows it to either reside within the genome of its host through lysogeny or enter into a lytic phase, during which it kills and lyses the cell to produce offspring. Lambda strains, mutated at specific sites, are unable to lysogenize cells; instead, they grow and enter the lytic cycle after superinfecting an already lysogenized cell.

Enterococcus faecalis – formerly classified as part of the group D Streptococcus system – is a Gram-positive, commensal bacterium inhabiting the gastrointestinal tracts of humans. Like other species in the genus Enterococcus, E. faecalis is found in healthy humans and can be used as a probiotic. The probiotic strains such as Symbioflor1 and EF-2001 are characterized by the lack of specific genes related to drug resistance and pathogenesis. As an opportunistic pathogen, E. faecalis can cause life-threatening infections, especially in the nosocomial (hospital) environment, where the naturally high levels of antibiotic resistance found in E. faecalis contribute to its pathogenicity. E. faecalis has been frequently found in reinfected, root canal-treated teeth in prevalence values ranging from 30% to 90% of the cases. Re-infected root canal-treated teeth are about nine times more likely to harbor E. faecalis than cases of primary infections.

The trp operon is a group of genes that are transcribed together, encoding the enzymes that produce the amino acid tryptophan in bacteria. The trp operon was first characterized in Escherichia coli, and it has since been discovered in many other bacteria. The operon is regulated so that, when tryptophan is present in the environment, the genes for tryptophan synthesis are repressed.
Rhodococcus fascians is a Gram positive bacterial phytopathogen that causes leafy gall disease. R. fascians is the only phytopathogenic member of the genus Rhodococcus; its host range includes both dicotyledonous and monocotyledonous hosts. Because it commonly afflicts tobacco (Nicotiana) plants, it is an agriculturally significant pathogen.
Addiction modules are toxin-antitoxin systems. Each consists of a pair of genes that specify two components: a stable toxin and an unstable antitoxin that interferes with the lethal action of the toxin. Found first in E. coli on low copy number plasmids, addiction modules are responsible for a process called the postsegregational killing effect. When bacteria lose these plasmid(s), the cured cells are selectively killed because the unstable antitoxin is degraded faster than the more stable toxin. The term "addiction" is used because the cell depends on the de novo synthesis of the antitoxin for cell survival. Thus, addiction modules are implicated in maintaining the stability of extrachromosomal elements.

The Tryptophan operon leader is an RNA element found at the 5′ of some bacterial tryptophan operons. The leader sequence can form two different structures known as the terminator and the anti-terminator, based on the Tryptophan amounts in the cell. The leader also codes for very short peptide sequence that is rich in tryptophan. The terminator structure is recognised as a termination signal for RNA polymerase and the operon is not transcribed. This structure forms when the cell has an excess of tryptophan and ribosome movement over the leader transcript is not impeded. When there is a deficiency of the charged tryptophanyl tRNA the ribosome translating the leader peptide stalls and the antiterminator structure can form. This allows RNA polymerase to transcribe the operon.
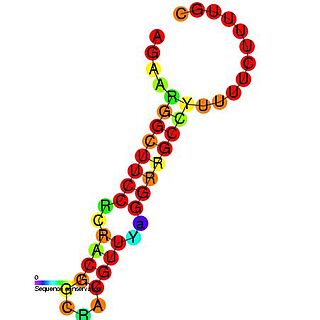
In molecular biology ctRNA is a plasmid encoded noncoding RNA that binds to the mRNA of repB and causes translational inhibition. ctRNA is encoded by plasmids and functions in rolling circle replication to maintain a low copy number. In Corynebacterium glutamicum, it achieves this by antisense pairing with the mRNA of RepB, a replication initiation protein. In Enterococcus faecium the plasmid pJB01 contains three open reading frames, copA, repB, and repC. The pJB01 ctRNA is coded on the opposite strand from the copA/repB intergenic region and partially overlaps an atypical ribosome binding site for repB.
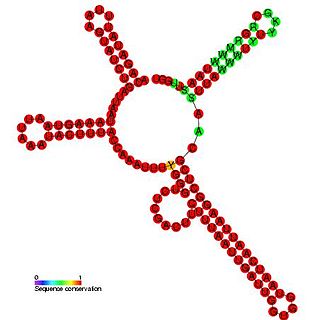
Plasmid RNAIII is a non-coding RNA found in bacterial plasmids including pIP501. RNAIII acts by transcriptional attenuation of the essential repR-mRNA. RNAIII is composed of four stem-loops with loops L3 and L4 that interact with the RNA target.

The Hfq protein encoded by the hfq gene was discovered in 1968 as an Escherichia coli host factor that was essential for replication of the bacteriophage Qβ. It is now clear that Hfq is an abundant bacterial RNA binding protein which has many important physiological roles that are usually mediated by interacting with Hfq binding sRNA.

The hok/sok system is a postsegregational killing mechanism employed by the R1 plasmid in Escherichia coli. It was the first type I toxin-antitoxin pair to be identified through characterisation of a plasmid-stabilising locus. It is a type I system because the toxin is neutralised by a complementary RNA, rather than a partnered protein.

Class II bacteriocins are a class of small peptides that inhibit the growth of various bacteria.
Post-transcriptional regulation is the control of gene expression at the RNA level. It occurs once the RNA polymerase has been attached to the gene's promoter and is synthesizing the nucleotide sequence. Therefore, as the name indicates, it occurs between the transcription phase and the translation phase of gene expression. These controls are critical for the regulation of many genes across human tissues. It also plays a big role in cell physiology, being implicated in pathologies such as cancer and neurodegenerative diseases.

cspA mRNA 5' UTR is the untranslated region of the cspA gene, which is important in the cold shock response in Enterobacteriales such as E. coli. The 5' UTR element acts as an RNA thermometer, regulating the expression of cspA in response to temperature. By regulating temperature, cspA proteins carry out the vital function of homeostasis.

FnrS RNA is a family of Hfq-binding small RNA whose expression is upregulated in response to anaerobic conditions. It is named FnrS because its expression is strongly dependent on fumarate and nitrate reductase regulator (FNR), a direct oxygen availability sensor.

A toxin-antitoxin system consists of a "toxin" and a corresponding "antitoxin", usually encoded by closely linked genes. The toxin is usually a protein while the antitoxin can be a protein or an RNA. Toxin-antitoxin systems are widely distributed in prokaryotes, and organisms often have them in multiple copies. When these systems are contained on plasmids – transferable genetic elements – they ensure that only the daughter cells that inherit the plasmid survive after cell division. If the plasmid is absent in a daughter cell, the unstable antitoxin is degraded and the stable toxic protein kills the new cell; this is known as 'post-segregational killing' (PSK).

The TxpA/RatA toxin-antitoxin system was first identified in Bacillus subtilis. It consists of a non-coding 222nt sRNA called RatA and a protein toxin named TxpA.

The par stability determinant is a 400 bp locus of the pAD1 plasmid which encodes a type I toxin-antitoxin system in Enterococcus faecalis. It was the first such plasmid addiction module to be found in gram-positive bacteria.
In molecular biology, the PyrG leader is a cis-regulatory RNA element found at the 5' of the PyrG mRNA. The PyrG gene encodes a CTP synthase, which is involved in pyrimidine biosynthesis. The PyrG leader regulates expression of PyrG, PyrG can form into two different hairpin structures, a terminator or an anti-terminator. Under low CTP conditions, guanine (G) residues are incorporated at a specific site within the PyrG leader, these allow base-pairing with a uracil (U)-rich region and the formation of an anti-terminator loop, this results in increased expression of PyrG. Under high CTP conditions the guanines are not added, the anti-terminator loop cannot form and instead a terminator loop is formed, preventing further PyrG expression.
Gelatinase biosynthesis-activating pheromone abbreviated as GBAP is a cyclic peptide produced by pathogenic bacteria such as Enterococcus faecalis. GAP is part of the quorum sensing system of certain bacteria where it positively regulates the expression of gelatinase and serine proteases that are under the control of the gelE-sprE operon.

The Enterococcus-1 RNA motif is a conserved RNA structure that was discovered by bioinformatics. Enterococcus-1 motif RNAs are found in bacteria of the genus Enterococcus.
















