| Autotransporter beta-domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | Autotransporter | ||||||||
| Pfam | PF03797 | ||||||||
| InterPro | IPR005546 | ||||||||
| PROSITE | PDOC51208 | ||||||||
| SCOP | 1uyn | ||||||||
| SUPERFAMILY | 1uyn | ||||||||
| TCDB | 1.B.12 | ||||||||
| OPM superfamily | 28 | ||||||||
| OPM protein | 1uyo | ||||||||
| |||||||||
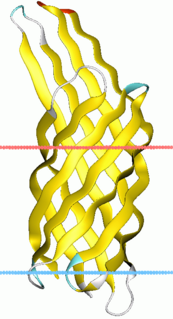
In molecular biology, an autotransporter domain is a structural domain found in some bacterial outer membrane proteins. The domain is always located at the C-terminal end of the protein and forms a beta-barrel structure. The barrel is oriented in the membrane such that the N-terminal portion of the protein, termed the passenger domain, is presented on the cell surface. These proteins are typically virulence factors, associated with infection or virulence in pathogenic bacteria.

A protein domain is a conserved part of a given protein sequence and (tertiary) structure that can evolve, function, and exist independently of the rest of the protein chain. Each domain forms a compact three-dimensional structure and often can be independently stable and folded. Many proteins consist of several structural domains. One domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. In general, domains vary in length from between about 50 amino acids up to 250 amino acids in length. The shortest domains, such as zinc fingers, are stabilized by metal ions or disulfide bridges. Domains often form functional units, such as the calcium-binding EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimeric proteins.

The bacterial outer membrane is found in gram-negative bacteria. Its composition is distinct from that of the inner cytoplasmic cell membrane - among other things, the outer leaflet of the outer membrane of many gram-negative bacteria includes a complex lipopolysaccharide whose lipid portion acts as an endotoxin - and in some bacteria such as E. coli it is linked to the cell's peptidoglycan by Braun's lipoprotein.
Contents
The name autotransporter derives from an initial understanding that the protein was self-sufficient in transporting the passenger domain through the outermembrane. This view has since been challenged by Benz and Schmidt. [2]
Secretion of polypeptide chains through the outer membrane of Gram-negative bacteria can occur via a number of different pathways. The type V(a), or autotransporter, secretion pathway constitutes the largest number of secreted virulence factors of any one of the seven known types of secretion in Gram-negative bacteria. This secretion pathway is exemplified by the prototypical IgA1 Protease of Neisseria gonorrhoeae. [3] The protein is directed to the inner membrane by a signal peptide transported across the inner membrane via the Sec machinery. Once in the periplasm, the autotransporter domain inserts into the outer membrane. The passenger domain is passed through the center of the autotransporter domain to be presented on the outside of the cell, however the mechanism by which this occurs remains unclear. [4]

Gram-negative bacteria are bacteria that do not retain the crystal violet stain used in the gram-staining method of bacterial differentiation. They are characterized by their cell envelopes, which are composed of a thin peptidoglycan cell wall sandwiched between an inner cytoplasmic cell membrane and a bacterial outer membrane.
A signal peptide is a short peptide present at the N-terminus of the majority of newly synthesized proteins that are destined towards the secretory pathway. These proteins include those that reside either inside certain organelles, secreted from the cell, or inserted into most cellular membranes. Although most type I membrane-bound proteins have signal peptides, the majority of type II and multi-spanning membrane-bound proteins are targeted to the secretory pathway by their first transmembrane domain, which biochemically resembles a signal sequence except that it is not cleaved.

The periplasm is a concentrated gel-like matrix in the space between the inner cytoplasmic membrane and the bacterial outer membrane called the periplasmic space in gram-negative bacteria. Using cryo-electron microscopy it has been found that a much smaller periplasmic space is also present in gram-positive bacteria.
The C-terminal translocator domain corresponds to an outer membrane beta-barrel domain. The N-terminal passenger domain is translocated across the membrane, and may or may not be cleaved from the translocator domain. [5] In those proteins where the cleavage is auto-catalytic, the peptidase domains belong to MEROPS peptidase families S6 and S8. Passenger domains structurally characterized to date have been shown to be dominated by a protein fold known as a beta helix, typified by pertactin. The folding of this domain is thought to be intrinsically linked to its method of outer membrane translocation.

A beta helix is a tandem protein repeat structure formed by the association of parallel beta strands in a helical pattern with either two or three faces. The beta helix is a type of solenoid protein domain. The structure is stabilized by inter-strand hydrogen bonds, protein-protein interactions, and sometimes bound metal ions. Both left- and right-handed beta helices have been identified. Double stranded beta-helices are also very common features of proteins and are generally synonymous with jelly roll folds.

In molecular biology, pertactin (PRN) is a highly immunogenic virulence factor of Bordetella pertussis, the bacterium that causes pertussis. Specifically, it is an outer membrane protein that promotes adhesion to tracheal epithelial cells. PRN is purified from Bordetella pertussis and is used for the vaccine production as one of the important components of acellular pertussis vaccine.