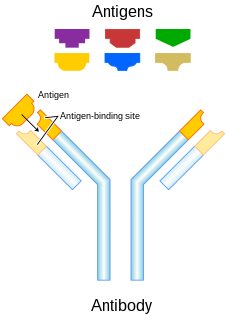
An antibody (Ab), also known as an immunoglobulin (Ig), is a large, Y-shaped protein produced mainly by plasma cells that is used by the immune system to neutralize pathogens such as pathogenic bacteria and viruses. The antibody recognizes a unique molecule of the pathogen, called an antigen, via the Fab's variable region. Each tip of the "Y" of an antibody contains a paratope that is specific for one particular epitope on an antigen, allowing these two structures to bind together with precision. Using this binding mechanism, an antibody can tag a microbe or an infected cell for attack by other parts of the immune system, or can neutralize its target directly. Depending on the antigen, the binding may impede the biological process causing the disease or may activate macrophages to destroy the foreign substance. The ability of an antibody to communicate with the other components of the immune system is mediated via its Fc region, which contains a conserved glycosylation site involved in these interactions. The production of antibodies is the main function of the humoral immune system.
Immunoglobulin M (IgM) is one of several forms of antibody that are produced by vertebrates. IgM is the largest antibody, and it is the first antibody to appear in the response to initial exposure to an antigen. In the case of humans and other mammals that have been studied, the spleen, where plasmablasts responsible for antibody production reside, is the major site of specific IgM production.

The immunoglobulin heavy chain (IgH) is the large polypeptide subunit of an antibody (immunoglobulin).
V(D)J recombination is the unique mechanism of genetic recombination that occurs only in developing lymphocytes during the early stages of T and B cell maturation. It involves somatic recombination, and results in the highly diverse repertoire of antibodies/immunoglobulins (Igs) and T cell receptors (TCRs) found on B cells and T cells, respectively. The process is a defining feature of the adaptive immune system.

The B-cell receptor or BCR is composed of immunoglobulin molecules that form a type 1 transmembrane receptor protein usually located on the outer surface of a lymphocyte type known as B cells. Through biochemical signaling and by physically acquiring antigens from the immune synapses, the BCR controls the activation of B-cell. B cells are able to gather and grab antigens by engaging biochemical modules for receptor clustering, cell spreading, generation of pulling forces, and receptor transport, which eventually culminates in endocytosis and antigen presentation. B-cells’ mechanical activity adheres to a pattern of negative and positive feedbacks that regulate the quantity of removed antigen by manipulating the dynamic of BCR-antigen bonds directly. Particularly, grouping and spreading increase the relation of antigen with BCR, thereby proving sensitivity and amplification. On the other hand, pulling forces delinks the antigen from the BCR, thus testing the quality of antigen binding.

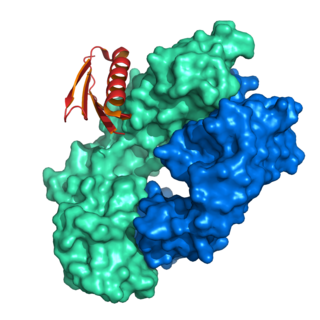
Protein A is a 42 kDa surface protein originally found in the cell wall of the bacteria Staphylococcus aureus. It is encoded by the spa gene and its regulation is controlled by DNA topology, cellular osmolarity, and a two-component system called ArlS-ArlR. It has found use in biochemical research because of its ability to bind immunoglobulins. It is composed of five homologous Ig-binding domains that fold into a three-helix bundle. Each domain is able to bind proteins from many mammalian species, most notably IgGs. It binds the heavy chain within the Fc region of most immunoglobulins and also within the Fab region in the case of the human VH3 family. Through these interactions in serum, where IgG molecules are bound in the wrong orientation, the bacteria disrupts opsonization and phagocytosis.

The immunoglobulin superfamily (IgSF) is a large protein superfamily of cell surface and soluble proteins that are involved in the recognition, binding, or adhesion processes of cells. Molecules are categorized as members of this superfamily based on shared structural features with immunoglobulins ; they all possess a domain known as an immunoglobulin domain or fold. Members of the IgSF include cell surface antigen receptors, co-receptors and co-stimulatory molecules of the immune system, molecules involved in antigen presentation to lymphocytes, cell adhesion molecules, certain cytokine receptors and intracellular muscle proteins. They are commonly associated with roles in the immune system. Otherwise, the sperm-specific protein IZUMO1, a member of the immunoglobulin superfamily, has also been identified as the only sperm membrane protein essential for sperm-egg fusion.

The fragment crystallizable region is the tail region of an antibody that interacts with cell surface receptors called Fc receptors and some proteins of the complement system. This property allows antibodies to activate the immune system. In IgG, IgA and IgD antibody isotypes, the Fc region is composed of two identical protein fragments, derived from the second and third constant domains of the antibody's two heavy chains; IgM and IgE Fc regions contain three heavy chain constant domains in each polypeptide chain. The Fc regions of IgGs bear a highly conserved N-glycosylation site. Glycosylation of the Fc fragment is essential for Fc receptor-mediated activity. The N-glycans attached to this site are predominantly core-fucosylated diantennary structures of the complex type. In addition, small amounts of these N-glycans also bear bisecting GlcNAc and α-2,6 linked sialic acid residues.
The complement component 1q is a protein complex involved in the complement system, which is part of the innate immune system. C1q together with C1r and C1s form the C1 complex.

Cytokine receptors are receptors that bind cytokines.

Immunoglobulin class switching, also known as isotype switching, isotypic commutation or class-switch recombination (CSR), is a biological mechanism that changes a B cell's production of immunoglobulin from one type to another, such as from the isotype IgM to the isotype IgG. During this process, the constant-region portion of the antibody heavy chain is changed, but the variable region of the heavy chain stays the same. Since the variable region does not change, class switching does not affect antigen specificity. Instead, the antibody retains affinity for the same antigens, but can interact with different effector molecules.

The immunoglobulin light chain is the small polypeptide subunit of an antibody (immunoglobulin).

Polymeric immunoglobulin receptor (pIgR) is a transmembrane protein that in humans is encoded by the PIGR gene. It is an Fc receptor which facilitates the transcytosis of the soluble polymeric isoforms of immunoglobulin A, and immune complexes. pIgRs are mainly located on the epithelial lining of mucosal surfaces of the gastrointestinal tract. The composition of the receptor is complex, including 6 immunoglobulin-like domains, a transmembrane region, and an intracellular domain. pIgR expression is under the strong regulation of cytokines, hormones, and pathogenic stimuli.
Immunoglobulin lambda-like polypeptide 1 is a protein that in humans is encoded by the IGLL1 gene. IGLL1 has also recently been designated CD179B.
Basal cell adhesion molecule, also known as Lutheran antigen, is a plasma membrane glycoprotein that in humans is encoded by the BCAM gene. BCAM has also recently been designated CD239.
Immunoglobulin heavy constant alpha 1 is a immunoglobulin gene with symbol IGHA1. It encodes a constant (C) segment of Immunoglobulin A heavy chain. Immunoglobulin A is an antibody that plays a critical role in immune function in the mucous membranes. IgA shows the same typical structure of other antibody classes, with two heavy chains and two light chains, and four distinct domains: one variable region, and three variable regions. As a major class of immunoglobulin in body secretions, IgA plays a role in defending against infection, as well as preventing the access of foreign antigens to the immunologic system.

Leukocyte immunoglobulin-like receptor subfamily A member 3 (LILR-A3) also known as CD85 antigen-like family member E (CD85e), immunoglobulin-like transcript 6 (ILT-6), and leukocyte immunoglobulin-like receptor 4 (LIR-4) is a protein that in humans is encoded by the LILRA3 gene located within the eukocyte receptor complex on chromosome 19q13.4. Unlike many of its family, LILRA3 lacks a transmembrane domain. The function of LILRA3 is currently unknown. however it is highly homologous to other LILR genes, and can bind human leukocyte antigen (HLA) class I. Therefore, if secreted, the LILRA3 might impair interactions of membrane-bound LILRs with their HLA ligands, thus modulating immune reactions and influencing susceptibility to disease.

Cluster of differentiation CD79A also known as B-cell antigen receptor complex-associated protein alpha chain and MB-1 membrane glycoprotein, is a protein that in humans is encoded by the CD79A gene.
In molecular biology, the domain B, refers to the immunoglobulin-binding domain found in the Staphylococcus aureus virulence factor protein A (SpA). Hence, it is abbreviated to SpAB.