
The Rossmann fold is a structural motif found in proteins that bind nucleotides, such as enzyme cofactors FAD, NAD+, and NADP+. This fold is composed of alternating beta strands and alpha helical segments where the beta strands are hydrogen bonded to each other forming an extended beta sheet and the alpha helices surround both faces of the sheet to produce a three-layered sandwich. The classical Rossmann fold contains six beta strands whereas Rossmann-like folds, sometimes referred to as Rossmannoid folds, contain only five strands. The initial beta-alpha-beta (bab) fold is the most conserved segment of the Rossmann fold. The motif is named after Michael Rossmann who first noticed this structural motif in the enzyme lactate dehydrogenase in 1970 and who later observed that this was a frequently occurring motif in nucleotide binding proteins.

Nitric oxide synthases (NOSs) are a family of enzymes catalyzing the production of nitric oxide (NO) from L-arginine. NO is an important cellular signaling molecule. It helps modulate vascular tone, insulin secretion, airway tone, and peristalsis, and is involved in angiogenesis and neural development. It may function as a retrograde neurotransmitter. Nitric oxide is mediated in mammals by the calcium-calmodulin controlled isoenzymes eNOS and nNOS. The inducible isoform, iNOS, involved in immune response, binds calmodulin at physiologically relevant concentrations, and produces NO as an immune defense mechanism, as NO is a free radical with an unpaired electron. It is the proximate cause of septic shock and may function in autoimmune disease.

Thioredoxin reductases are the only known enzymes to reduce thioredoxin (Trx). Two classes of thioredoxin reductase have been identified: one class in bacteria and some eukaryotes and one in animals. Both classes are flavoproteins which function as homodimers. Each monomer contains a FAD prosthetic group, a NADPH binding domain, and an active site containing a redox-active disulfide bond.
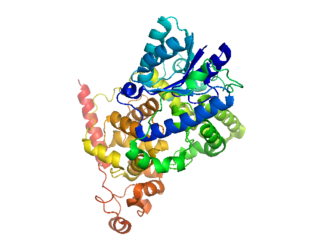
Cryptochromes are a class of flavoproteins that are sensitive to blue light. They are found in plants and animals. Cryptochromes are involved in the circadian rhythms of plants and animals, and possibly also in the sensing of magnetic fields in a number of species. The name cryptochrome was proposed as a portmanteau combining the cryptic nature of the photoreceptor, and the cryptogamic organisms on which many blue-light studies were carried out.

Flavoproteins are proteins that contain a nucleic acid derivative of riboflavin: the flavin adenine dinucleotide (FAD) or flavin mononucleotide (FMN).
Acyl-CoA dehydrogenases (ACADs) are a class of enzymes that function to catalyze the initial step in each cycle of fatty acid β-oxidation in the mitochondria of cells. Their action results in the introduction of a trans double-bond between C2 (α) and C3 (β) of the acyl-CoA thioester substrate. Flavin adenine dinucleotide (FAD) is a required co-factor in addition to the presence of an active site glutamate in order for the enzyme to function.

The acetolactate synthase (ALS) enzyme is a protein found in plants and micro-organisms. ALS catalyzes the first step in the synthesis of the branched-chain amino acids.

Glutaryl-CoA dehydrogenase (GCDH) is an enzyme encoded by the GCDH gene on chromosome 19. The protein belongs to the acyl-CoA dehydrogenase family (ACD). It catalyzes the oxidative decarboxylation of glutaryl-CoA to crotonyl-CoA and carbon dioxide in the degradative pathway of L-lysine, L-hydroxylysine, and L-tryptophan metabolism. It uses electron transfer flavoprotein as its electron acceptor. The enzyme exists in the mitochondrial matrix as a homotetramer of 45-kD subunits. Mutations in this gene result in the metabolic disorder glutaric aciduria type 1, which is also known as glutaric acidemia type I. Alternative splicing of this gene results in multiple transcript variants.
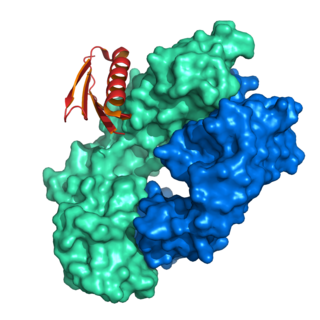
Protein L was first isolated from the surface of bacterial species Peptostreptococcus magnus and was found to bind immunoglobulins through L chain interaction, from which the name was suggested. It consists of 719 amino acid residues. The molecular weight of Protein L purified from the cell walls of Peptostreptoccus magnus was first estimated as 95kD by SDS-PAGE in the presence of reducing agent 2-mercaptoethanol, while the molecular weight was determined to 76kD by gel chromotography in the presence of 6 M guanidine HCl. Protein L does not contain any interchain disulfide loops, nor does it consist of disulfide-linked subunits. It is an acidic molecule with a pI of 4.0. Unlike Protein A and Protein G, which bind to the Fc region of immunoglobulins (antibodies), Protein L binds antibodies through light chain interactions. Since no part of the heavy chain is involved in the binding interaction, Protein L binds a wider range of antibody classes than Protein A or G. Protein L binds to representatives of all antibody classes, including IgG, IgM, IgA, IgE and IgD. Single chain variable fragments (scFv) and Fab fragments also bind to Protein L.
In enzymology, a ferredoxin-NADP+ reductase (EC 1.18.1.2) abbreviated FNR, is an enzyme that catalyzes the chemical reaction

The aldo-keto reductase family is a family of proteins that are subdivided into 16 categories; these include a number of related monomeric NADPH-dependent oxidoreductases, such as aldehyde reductase, aldose reductase, prostaglandin F synthase, xylose reductase, rho crystallin, and many others.
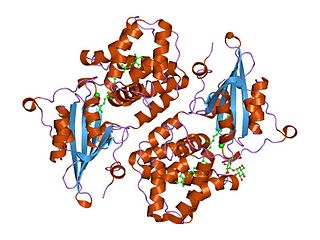
In molecular biology the orange carotenoid N-terminal domain is a protein domain found predominantly at the N-terminus of the Orange carotenoid protein (OCP), and is involved in non-covalent binding of a carotenoid chromophore. It is unique for being present in soluble proteins, whereas the vast majority of domains capable of binding carotenoids are intrinsic membrane proteins. Thus far, it has exclusively been found in cyanobacteria, among which it is widespread. The domain also exists on its own, in uncharacterized cyanobacterial proteins referred to as "Red Carotenoid Protein" (RCP). The domain adopts an alpha-helical structure consisting of two four-helix bundles.
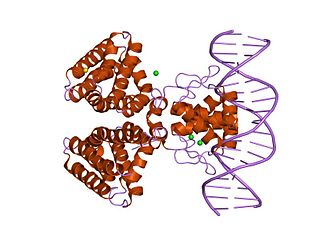
In molecular biology, the fatty acid metabolism regulator protein FadR, is a bacterial transcription factor.

In molecular biology, the GntR-like bacterial transcription factors are a family of transcription factors.
A Light-oxygen-voltage-sensing domain is a protein sensor used by a large variety of higher plants, microalgae, fungi and bacteria to sense environmental conditions. In higher plants, they are used to control phototropism, chloroplast relocation, and stomatal opening, whereas in fungal organisms, they are used for adjusting the circadian temporal organization of the cells to the daily and seasonal periods.
White Collar-1 (wc-1) is a gene in Neurospora crassa encoding the protein WC-1. WC-1 has two separate roles in the cell. First, it is the primary photoreceptor for Neurospora and the founding member of the class of principle blue light photoreceptors in all of the fungi. Second, it is necessary for regulating circadian rhythms in FRQ. It is a key component of a circadian molecular pathway that regulates many behavioral activities, including conidiation. WC-1 and WC-2, an interacting partner of WC-1, comprise the White Collar Complex (WCC) that is involved in the Neurospora circadian clock. WCC is a complex of nuclear transcription factor proteins, and contains transcriptional activation domains, PAS domains, and zinc finger DNA-binding domains (GATA). WC-1 and WC-2 heterodimerize through their PAS domains to form the White Collar Complex (WCC).

A FMN-binding fluorescent protein is a small, oxygen-independent fluorescent protein that binds flavin mononucleotide (FMN) as a chromophore.
The white collar--2 (wc-2) gene in Neurospora crassa encodes the protein White Collar-2 (WC-2). WC-2 is a GATA transcription factor necessary for blue light photoreception and for regulating circadian rhythms in Neurospora. In both contexts, WC-2 binds to its non-redundant counterpart White Collar-1 (WC-1) through PAS domains to form the White Collar Complex (WCC), an active transcription factor.

















