Metabolism is the set of life-sustaining chemical reactions in organisms. The three main functions of metabolism are: the conversion of the energy in food to energy available to run cellular processes; the conversion of food to building blocks of proteins, lipids, nucleic acids, and some carbohydrates; and the elimination of metabolic wastes. These enzyme-catalyzed reactions allow organisms to grow and reproduce, maintain their structures, and respond to their environments. The word metabolism can also refer to the sum of all chemical reactions that occur in living organisms, including digestion and the transportation of substances into and between different cells, in which case the above described set of reactions within the cells is called intermediary metabolism.
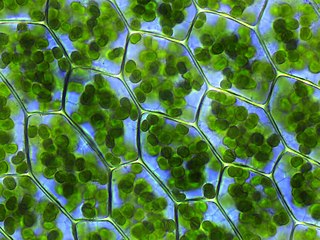
A plastid is a membrane-bound organelle found in the cells of plants, algae, and some other eukaryotic organisms. They are considered to be intracellular endosymbiotic cyanobacteria.

The mevalonate pathway, also known as the isoprenoid pathway or HMG-CoA reductase pathway is an essential metabolic pathway present in eukaryotes, archaea, and some bacteria. The pathway produces two five-carbon building blocks called isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP), which are used to make isoprenoids, a diverse class of over 30,000 biomolecules such as cholesterol, vitamin K, coenzyme Q10, and all steroid hormones.
Metabolic network modelling, also known as metabolic network reconstruction or metabolic pathway analysis, allows for an in-depth insight into the molecular mechanisms of a particular organism. In particular, these models correlate the genome with molecular physiology. A reconstruction breaks down metabolic pathways into their respective reactions and enzymes, and analyzes them within the perspective of the entire network. In simplified terms, a reconstruction collects all of the relevant metabolic information of an organism and compiles it in a mathematical model. Validation and analysis of reconstructions can allow identification of key features of metabolism such as growth yield, resource distribution, network robustness, and gene essentiality. This knowledge can then be applied to create novel biotechnology.
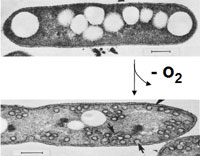
An apicoplast is a derived non-photosynthetic plastid found in most Apicomplexa, including Toxoplasma gondii, and Plasmodium falciparum and other Plasmodium spp., but not in others such as Cryptosporidium. It originated from algae through secondary endosymbiosis; there is debate as to whether this was a green or red alga. The apicoplast is surrounded by four membranes within the outermost part of the endomembrane system. The apicoplast hosts important metabolic pathways like fatty acid synthesis, isoprenoid precursor synthesis and parts of the heme biosynthetic pathway.
The non-mevalonate pathway—also appearing as the mevalonate-independent pathway and the 2-C-methyl-D-erythritol 4-phosphate/1-deoxy-D-xylulose 5-phosphate (MEP/DOXP) pathway—is an alternative metabolic pathway for the biosynthesis of the isoprenoid precursors isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP). The currently preferred name for this pathway is the MEP pathway, since MEP is the first committed metabolite on the route to IPP.

Mevalonate kinase is an enzyme that in humans is encoded by the MVK gene. Mevalonate kinases are found in a wide variety of organisms from bacteria to mammals. This enzyme catalyzes the following reaction:

Apolipoprotein A-II is a protein that in humans is encoded by the APOA2 gene. It is the second most abundant protein of the high density lipoprotein particles. The protein is found in plasma as a monomer, homodimer, or heterodimer with apolipoprotein D. ApoA-II regulates many steps in HDL metabolism, and its role in coronary heart disease is unclear. Remarkably, defects in this gene may result in apolipoprotein A-II deficiency or hypercholesterolemia.
The enzyme phosphoketolase(EC 4.1.2.9) catalyzes the chemical reactions

Archaea is a domain of single-celled organisms. These microorganisms lack cell nuclei and are therefore prokaryotic. Archaea were initially classified as bacteria, receiving the name archaebacteria, but this term has fallen out of use.

Nannochloropsis is a genus of algae comprising six known species. The genus in the current taxonomic classification was first termed by Hibberd (1981). The species have mostly been known from the marine environment but also occur in fresh and brackish water. All of the species are small, nonmotile spheres which do not express any distinct morphological features that can be distinguished by either light or electron microscopy. The characterisation is mostly done by rbcL gene and 18S rRNA sequence analysis.
Tryptophan-rich sensory proteins (TspO) are a family of proteins that are involved in transmembrane signalling. In either prokaryotes or mitochondria they are localized to the outer membrane, and have been shown to bind and transport dicarboxylic tetrapyrrole intermediates of the haem biosynthetic pathway. They are associated with the major outer membrane porins and with the voltage-dependent anion channel.
Rhodovulum sulfidophilum is a gram-negative purple nonsulfur bacteria. The cells are rod-shaped, and range in size from 0.6 to 0.9 μm wide and 0.9 to 2.0 μm long, and have a polar flagella. These cells reproduce asexually by binary fission. This bacterium can grow anaerobically when light is present, or aerobically (chemoheterotrophic) under dark conditions. It contains the photosynthetic pigments bacteriochlorophyll a and of carotenoids.
Chlorobaculum tepidum, previously known as Chlorobium tepidum, is an anaerobic, thermophilic green sulfur bacteria first isolated from New Zealand. Its cells are gram-negative and non-motile rods of variable length. They contain chlorosomes and bacteriochlorophyll a and c.

Antonius Suwanto, is an Indonesian biologist and professor, known for discovering two circular chromosomes or plasmids in Rhodobacter with Kaplan S in 1989.
Rhodobacter capsulatus is a species of purple bacteria, a group of bacteria that can obtain energy through photosynthesis. Its name is derived from the Latin adjective "capsulatus", itself derived Latin noun "capsula", and the associated Latin suffix for masculine nouns, "-atus".
Robert L. Last is a plant biochemical genomicist who studies metabolic processes that protect plants from the environment and produce products important for animal and human nutrition. His research has covered (1) production and breakdown of essential amino acids, (2) the synthesis and protective roles of Vitamin C and Vitamin E (tocopherols) as well as identification of mechanisms that protect photosystem II from damage, and (3) synthesis and biological functions of plant protective specialized metabolites. Four central questions are: (i) how are leaf and seed amino acids levels regulated, (ii.) what mechanisms protect and repair photosystem II from stress-induced damage, (iii.) how do plants produce protective metabolites in their glandular secreting trichomes (iv.) and what are the evolutionary mechanisms that contribute to the tremendous diversity of specialized metabolites that protect plants from insects and pathogens and are used as therapeutic agents.
Auxiliary metabolic genes (AMGs) are found in many bacteriophages but originated in bacterial cells. AMGs modulate host cell metabolism during infection so that the phage can replicate more efficiently. For instance, bacteriophages that infect the abundant marine cyanobacteria Synechococcus and Prochlorococcus (cyanophages) carry AMGs that have been acquired from their immediate host as well as more distantly-related bacteria. Cyanophage AMGs support a variety of functions including photosynthesis, carbon metabolism, nucleic acid synthesis and metabolism. AMGs also have broader ecological impacts beyond their host including their influence on biogeochemical cycling.

Chlorophyllide a and Chlorophyllide b are the biosynthetic precursors of chlorophyll a and chlorophyll b respectively. Their propionic acid groups are converted to phytyl esters by the enzyme chlorophyll synthase in the final step of the pathway. Thus the main interest in these chemical compounds has been in the study of chlorophyll biosynthesis in plants, algae and cyanobacteria. Chlorophyllide a is also an intermediate in the biosynthesis of bacteriochlorophylls.

Chlorophyllide a reductase (EC 1.3.7.15), also known as COR, is an enzyme with systematic name bacteriochlorophyllide-a:ferredoxin 7,8-oxidoreductase. It catalyses the following chemical reaction