
A spirochaete or spirochete is a member of the phylum Spirochaetota, which contains distinctive diderm (double-membrane) Gram-negative bacteria, most of which have long, helically coiled cells. Spirochaetes are chemoheterotrophic in nature, with lengths between 3 and 500 μm and diameters around 0.09 to at least 3 μm.

The green sulfur bacteria are a phylum, Chlorobiota, of obligately anaerobic photoautotrophic bacteria that metabolize sulfur.
The Aquificota phylum is a diverse collection of bacteria that live in harsh environmental settings. The name Aquificota was given to this phylum based on an early genus identified within this group, Aquifex, which is able to produce water by oxidizing hydrogen. They have been found in springs, pools, and oceans. They are autotrophs, and are the primary carbon fixers in their environments. These bacteria are Gram-negative, non-spore-forming rods. They are true bacteria as opposed to the other inhabitants of extreme environments, the Archaea.
The Chloroflexia are a class of bacteria in the phylum Chloroflexota. Chloroflexia are typically filamentous, and can move about through bacterial gliding. It is named after the order Chloroflexales.

The phylum Bacteroidota is composed of three large classes of Gram-negative, nonsporeforming, anaerobic or aerobic, and rod-shaped bacteria that are widely distributed in the environment, including in soil, sediments, and sea water, as well as in the guts and on the skin of animals.
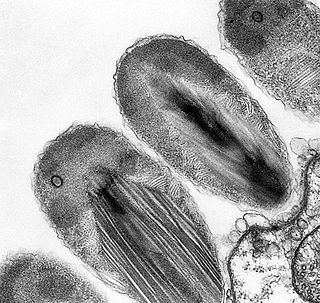
Verrucomicrobiota is a phylum of Gram-negative bacteria that contains only a few described species. The species identified have been isolated from fresh water, marine and soil environments and human faeces. A number of as-yet uncultivated species have been identified in association with eukaryotic hosts including extrusive explosive ectosymbionts of protists and endosymbionts of nematodes from genus Xiphinema, residing in their gametes. The verrucomicrobial bacterium Akkermansia muciniphila is a human intestinal symbiotic bacterium that is considered as a promising probiotic.

The Chlamydiota are a bacterial phylum and class whose members are remarkably diverse, including pathogens of humans and animals, symbionts of ubiquitous protozoa, and marine sediment forms not yet well understood. All of the Chlamydiota that humans have known about for many decades are obligate intracellular bacteria; in 2020 many additional Chlamydiota were discovered in ocean-floor environments, and it is not yet known whether they all have hosts. Historically it was believed that all Chlamydiota had a peptidoglycan-free cell wall, but studies in the 2010s demonstrated a detectable presence of peptidoglycan, as well as other important proteins.

The Campylobacterales are an order of Campylobacterota which make up the epsilon subdivision, together with the small family Nautiliaceae. They are Gram-negative. Most of the species are microaerophilic.
The Thermotogota are a phylum of the domain Bacteria. The phylum contains a single class, Thermotogae. The phylum Thermotogota is composed of Gram-negative staining, anaerobic, and mostly thermophilic and hyperthermophilic bacteria. It is the sole phylum in the kingdom Thermotogati.

The Bifidobacteriaceae are the only family of bacteria in the order Bifidobacteriales. According to the 16S rRNA-based LTP release 106 published by 'The All-Species Living Tree' Project, the order Bifidobacteriales is a clade nested within the suborder Micrococcineae, also the genus Bifidobacterium is paraphyletic to the other genera within the family, i.e. the other genera are nested within Bifidobacterium.
Fibrobacterota is a small bacterial phylum which includes many of the major rumen bacteria, allowing for the degradation of plant-based cellulose in ruminant animals. Members of this phylum were categorized in other phyla. The genus Fibrobacter was removed from the genus Bacteroides in 1988.

Streptomycetaceae is a family of the class Actinomycetota, making up the monotypic order Streptomycetales. It includes the important genus Streptomyces. This was the original source of many antibiotics, namely streptomycin, the first antibiotic against tuberculosis.

The PVC superphylum is a superphylum of bacteria named after its three important members, Planctomycetota, Verrucomicrobiota, and Chlamydiota. Cavalier-Smith postulated that the PVC bacteria probably lost or reduced their peptidoglycan cell wall twice. It has been hypothesised that a member of the PVC clade might have been the host cell in the endosymbiotic event that gave rise to the first proto-eukaryotic cell.

The order Flavobacteriales comprises several families of environmental bacteria.
Fibrobacter succinogenes is a cellulolytic bacterium species in the genus Fibrobacter. It is present in the rumen of cattle. F. succinogenes is a gram negative, rod-shaped, obligate anaerobe that is a major contributor to cellulose digestion. Since its discovery in the 1950s, it has been studied for its role in herbivore digestion and cellulose fermentation, which can be utilized in biofuel production.
The Synergistota is a phylum of anaerobic bacteria that show Gram-negative staining and have rod/vibrioid cell shape. Although Synergistota have a diderm cell envelope, the genes for various proteins involved in lipopolysaccharides biosynthesis have not yet been detected in Synergistota, indicating that they may have an atypical outer cell envelope. The Synergistota inhabit a majority of anaerobic environments including animal gastrointestinal tracts, soil, oil wells, and wastewater treatment plants and they are also present in sites of human diseases such as cysts, abscesses, and areas of periodontal disease. Due to their presence at illness related sites, the Synergistota are suggested to be opportunistic pathogens but they can also be found in healthy individuals in the microbiome of the umbilicus and in normal vaginal flora. Species within this phylum have also been implicated in periodontal disease, gastrointestinal infections and soft tissue infections. Other species from this phylum have been identified as significant contributors in the degradation of sludge for production of biogas in anaerobic digesters and are potential candidates for use in renewable energy production through their production of hydrogen gas. All of the known Synergistota species and genera are presently part of a single class (Synergistia), order (Synergistiales), and family (Synergistaceae).

The FCB group is a superphylum of bacteria named after the main member phyla Fibrobacterota, Chlorobiota, and Bacteroidota. The members are considered to form a clade due to a number of conserved signature indels.
Adlercreutzia is a genus in the phylum Actinomycetota (Bacteria).
Conserved signature inserts and deletions (CSIs) in protein sequences provide an important category of molecular markers for understanding phylogenetic relationships. CSIs, brought about by rare genetic changes, provide useful phylogenetic markers that are generally of defined size and they are flanked on both sides by conserved regions to ensure their reliability. While indels can be arbitrary inserts or deletions, CSIs are defined as only those protein indels that are present within conserved regions of the protein.
Thermaceae is a family of bacteria belonging to the phylum Deinococcota. It is the only family in the order Thermales. They are particularly resistant to heat, and live in the benthic zone of the Gulf of Mexico.









