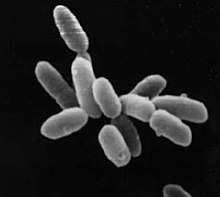
A halophile is an extremophile that thrives in high salt concentrations. In chemical terms, halophile refers to a Lewis acidic species that has some ability to extract halides from other chemical species.

The phylum Bacteroidota is composed of three large classes of Gram-negative, nonsporeforming, anaerobic or aerobic, and rod-shaped bacteria that are widely distributed in the environment, including in soil, sediments, and sea water, as well as in the guts and on the skin of animals.

Sulfolobus is a genus of microorganism in the family Sulfolobaceae. It belongs to the archaea domain.
Halobacteriaceae is a family in the order Halobacteriales and the domain Archaea. Halobacteriaceae represent a large part of halophilic Archaea, along with members in two other methanogenic families, Methanosarcinaceae and Methanocalculaceae. The family consists of many diverse genera that can survive extreme environmental niches. Most commonly, Halobacteriaceae are found in hypersaline lakes and can even tolerate sites polluted by heavy metals. They include neutrophiles, acidophiles, alkaliphiles, and there have even been psychrotolerant species discovered. Some members have been known to live aerobically, as well as anaerobically, and they come in many different morphologies. These diverse morphologies include rods in genus Halobacterium, cocci in Halococcus, flattened discs or cups in Haloferax, and other shapes ranging from flattened triangles in Haloarcula to squares in Haloquadratum, and Natronorubrum. Most species of Halobacteriaceae are best known for their high salt tolerance and red-pink pigmented members, but there are also non-pigmented species and those that require moderate salt conditions. Some species of Halobacteriaceae have been shown to exhibit phosphorus solubilizing activities that contribute to phosphorus cycling in hypersaline environments. Techniques such as 16S rRNA analysis and DNA–DNA hybridization have been major contributors to taxonomic classification in Halobacteriaceae, partly due to the difficulty in culturing halophilic Archaea.

Halobacteriales are an order of the Halobacteria, found in water saturated or nearly saturated with salt. They are also called halophiles, though this name is also used for other organisms which live in somewhat less concentrated salt water. They are common in most environments where large amounts of salt, moisture, and organic material are available. Large blooms appear reddish, from the pigment bacteriorhodopsin. This pigment is used to absorb light, which provides energy to create ATP. Halobacteria also possess a second pigment, halorhodopsin, which pumps in chloride ions in response to photons, creating a voltage gradient and assisting in the production of energy from light. The process is unrelated to other forms of photosynthesis involving electron transport; however, and halobacteria are incapable of fixing carbon from carbon dioxide.

Haloarchaea are a class of prokaryotic archaea under the phylum Euryarchaeota, found in water saturated or nearly saturated with salt. 'Halobacteria' are now recognized as archaea rather than bacteria and are one of the largest groups or archaea. The name 'halobacteria' was assigned to this group of organisms before the existence of the domain Archaea was realized, and while valid according to taxonomic rules, should be updated. Halophilic archaea are generally referred to as haloarchaea to distinguish them from halophilic bacteria.
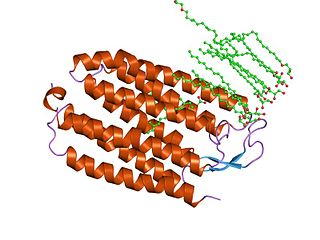
Halorhodopsin is a seven-transmembrane retinylidene protein from microbial rhodopsin family. It is a chloride-specific light-activated ion pump found in archaea known as halobacteria. It is activated by green light wavelengths of approximately 578 nm. Halorhodopsin also shares sequence similarity to channelrhodopsin, a light-gated ion channel.

Sulfolobales is an order of archaeans in the class Thermoprotei.
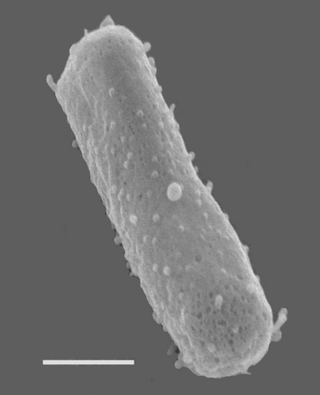
Halobacterium salinarum, formerly known as Halobacterium cutirubrum or Halobacterium halobium, is an extremely halophilic marine obligate aerobic archaeon. Despite its name, this is not a bacterium, but a member of the domain Archaea. It is found in salted fish, hides, hypersaline lakes, and salterns. As these salterns reach the minimum salinity limits for extreme halophiles, their waters become purple or reddish color due to the high densities of halophilic Archaea. H. salinarum has also been found in high-salt food such as salt pork, marine fish, and sausages. The ability of H. salinarum to live at such high salt concentrations has led to its classification as an extremophile.
Halorubrum is a genus in the family Halorubraceae. Halorubrum species are usually halophilic and can be found in waters with high salt concentration such as the Dead Sea or Lake Zabuye.
In taxonomy, Natrialba is a genus of the Natrialbaceae. The genus consists of many diverse species that can survive extreme environmental niches, especially they are capable to live in the waters saturated or nearly saturated with salt (halophiles). They have certain adaptations to live within their salty environments. For example, their cellular machinery is adapted to high salt concentrations by having charged amino acids on their surfaces, allowing the cell to keep its water molecules around these components. The osmotic pressure and these amino acids help to control the amount of salt within the cell.

A pink lake is a lake that has a red or pink colour. This is often caused by the presence of salt-tolerant algae that produces carotenoids, such as Dunaliella salina, usually in conjunction with specific bacteria and archaea, which may vary from lake to lake. The most common archaeon is Halobacterium salinarum.

Haloferax volcanii is a species of organism in the genus Haloferax in the Archaea.
Halobacterium noricense is a halophilic, rod-shaped microorganism that thrives in environments with salt levels near saturation. Despite the implication of the name, Halobacterium is actually a genus of archaea, not bacteria. H. noricense can be isolated from environments with high salinity such as the Dead Sea and the Great Salt Lake in Utah. Members of the Halobacterium genus are excellent model organisms for DNA replication and transcription due to the stability of their proteins and polymerases when exposed to high temperatures. To be classified in the genus Halobacterium, a microorganism must exhibit a membrane composition consisting of ether-linked phosphoglycerides and glycolipids.

Microbial rhodopsins, also known as bacterial rhodopsins, are retinal-binding proteins that provide light-dependent ion transport and sensory functions in halophilic and other bacteria. They are integral membrane proteins with seven transmembrane helices, the last of which contains the attachment point for retinal. Most microbial rhodopsins pump inwards, however "mirror rhodopsins" which function outwards. have been discovered.

Gas vesicles, also known as gas vacuoles, are nanocompartments in certain prokaryotic organisms, which help in buoyancy. Gas vesicles are composed entirely of protein; no lipids or carbohydrates have been detected.

Shiladitya DasSarma is a molecular biologist well-known for contributions to the biology of halophilic and extremophilic microorganisms. He is a Professor in the University of Maryland Baltimore. He earned a PhD degree in biochemistry from the Massachusetts Institute of Technology and a BS degree in chemistry from Indiana University Bloomington. Prior to taking a faculty position, he conducted research at the Massachusetts General Hospital, Harvard Medical School, and Pasteur Institute, Paris.

Halorubrum lacusprofundi is a rod-shaped, halophilic Archaeon in the family of Halorubraceae. It was first isolated from Deep Lake in Antarctica in the 1980s.

Archaerhodopsin proteins are a family of retinal-containing photoreceptors found in the archaea genera Halobacterium and Halorubrum. Like the homologous bacteriorhodopsin (bR) protein, archaerhodopsins harvest energy from sunlight to pump H+ ions out of the cell, establishing a proton motive force that is used for ATP synthesis. They have some structural similarities to the mammalian G protein-coupled receptor protein rhodopsin, but are not true homologs.
Halorubrum kocurii is a halophilic archaean belonging to the genus Halorubrum. This genus contains a total of thirty-seven different species, all of which thrive in high-salinity environments. Archaea belonging to this genus are typically found in hypersaline environments due to their halophilic nature, specifically in solar salterns. Halorubrum kocurii is a rod-shaped, Gram-negative archaeon. Different from its closest relatives, Halorubrum kocurii is non-motile and contains no flagella or cilia. This archaeon thrives at high pH levels, high salt concentrations, and moderate temperatures. It has a number of close relatives, including Halorubrum aidingense, Halorubrum lacusprofundi, and more.