| Nanoarchaeota | |
|---|---|
 | |
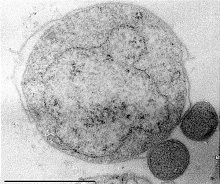
| Nanoarcheotum Nanopusillus acidilobi attached to Acidilobus . | |
| Scientific classification | |
| Domain: | Archaea |
| Kingdom: | Nanobdellati |
| Phylum: | Nanobdellota |
| Class: | Nanobdellia Kato et al. 2022 |
| Orders | |
| |
| Synonyms | |
| |
Nanoarchaeota (Greek, "dwarf or tiny ancient one") is a proposed phylum (Candidatus Nanoarchaeota) in the domain Archaea [1] that currently has only one representative, Nanoarchaeum equitans , which was discovered in a submarine hydrothermal vent and first described in 2002. [2]