| BSD | |||||||||
|---|---|---|---|---|---|---|---|---|---|
solution structure of the bsd domain of human tfiih basal transcription factor complex p62 subunit | |||||||||
| Identifiers | |||||||||
| Symbol | BSD | ||||||||
| Pfam | PF03909 | ||||||||
| InterPro | IPR005607 | ||||||||
| |||||||||
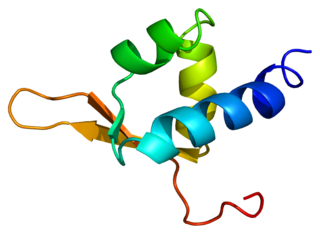
In molecular biology, the BSD domain is an approximately 60-amino-acid-long protein domain named after the BTF2-like transcription factors, synapse-associated proteins and DOS2-like proteins in which it is found. It is also found in several hypothetical proteins. The BSD domain occurs in one or two copies in a variety of species ranging from primal protozoan to human. It can be found associated with other domains such as the BTB domain or the U-box in multidomain proteins. The function of the BSD domain is as yet unknown. [1]

A protein domain is a conserved part of a given protein sequence and (tertiary) structure that can evolve, function, and exist independently of the rest of the protein chain. Each domain forms a compact three-dimensional structure and often can be independently stable and folded. Many proteins consist of several structural domains. One domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. In general, domains vary in length from between about 50 amino acids up to 250 amino acids in length. The shortest domains, such as zinc fingers, are stabilized by metal ions or disulfide bridges. Domains often form functional units, such as the calcium-binding EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimeric proteins.

In the nervous system, a synapse is a structure that permits a neuron to pass an electrical or chemical signal to another neuron or to the target effector cell.

In taxonomy, Homo sapiens is the only extant human species. The name is Latin for "wise man" and was introduced in 1758 by Carl Linnaeus.
Secondary structure prediction indicates the presence of three predicted alpha helices, which probably form a three-helical bundle in small |domains. The third predicted helix contains neighbouring phenylalanine and tryptophan residues – less common amino acids that are invariant in all the BSD domains identified and that are the most striking sequence features of the domain. [1]
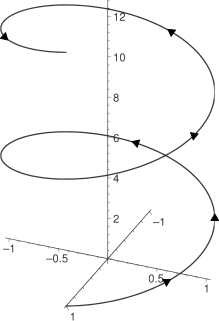
A helix, plural helixes or helices, is a type of smooth space curve, i.e. a curve in three-dimensional space. It has the property that the tangent line at any point makes a constant angle with a fixed line called the axis. Examples of helices are coil springs and the handrails of spiral staircases. A "filled-in" helix – for example, a "spiral" (helical) ramp – is called a helicoid. Helices are important in biology, as the DNA molecule is formed as two intertwined helices, and many proteins have helical substructures, known as alpha helices. The word helix comes from the Greek word ἕλιξ, "twisted, curved".

Phenylalanine is an essential α-amino acid with the formula C
9H
11NO
2. It can be viewed as a benzyl group substituted for the methyl group of alanine, or a phenyl group in place of a terminal hydrogen of alanine. This essential amino acid is classified as neutral, and nonpolar because of the inert and hydrophobic nature of the benzyl side chain. The L-isomer is used to biochemically form proteins, coded for by DNA. Phenylalanine is a precursor for tyrosine, the monoamine neurotransmitters dopamine, norepinephrine (noradrenaline), and epinephrine (adrenaline), and the skin pigment melanin. It is encoded by the codons UUU and UUC.

Tryptophan is an α-amino acid that is used in the biosynthesis of proteins. Tryptophan contains an α-amino group, an α-carboxylic acid group, and a side chain indole, making it a non-polar aromatic amino acid. It is essential in humans, meaning the body cannot synthesize it: it must be obtained from the diet. Tryptophan is also a precursor to the neurotransmitter serotonin, the hormone melatonin and vitamin B3. It is encoded by the codon UGG.
Some proteins known to contain one or two BSD domains are listed below:
- Mammalian TFIIH basal transcription factor complex p62 subunit (GTF2H1).
- Yeast RNA polymerase II transcription factor B 73 kDa subunit (TFB1), the homologue of BTF2.
- Yeast DOS2 protein, involved in single-copy DNA replication and ubiquitination.
- Drosophila synapse-associated protein SAP47.
- Mammalian SYAP1.
- Various Arabidopsis thaliana (mouse-ear cress) hypothetical proteins.

In molecular biology, a transcription factor (TF) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are expressed in the right cell at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are up to 2600 TFs in the human genome.

A protein complex or multiprotein complex is a group of two or more associated polypeptide chains. Different polypeptide chains may have different functions. This is distinct from a multienzyme complex, in which multiple catalytic domains are found in a single polypeptide chain.

General transcription factor IIH subunit 1 is a protein that in humans is encoded by the GTF2H1 gene.