Virus classification is the process of naming viruses and placing them into a taxonomic system similar to the classification systems used for cellular organisms.

A prophage is a bacteriophage genome that is integrated into the circular bacterial chromosome or exists as an extrachromosomal plasmid within the bacterial cell. Integration of prophages into the bacterial host is the characteristic step of the lysogenic cycle of temperate phages. Prophages remain latent in the genome through multiple cell divisions until activation by an external factor, such as UV light, leading to production of new phage particles that will lyse the cell and spread. As ubiquitous mobile genetic elements, prophages play important roles in bacterial genetics and evolution, such as in the acquisition of virulence factors.

Hepadnaviridae is a family of viruses. Humans, apes, and birds serve as natural hosts. There are currently 18 species in this family, divided among 5 genera. Its best-known member is hepatitis B virus. Diseases associated with this family include: liver infections, such as hepatitis, hepatocellular carcinomas, and cirrhosis. It is the sole accepted family in the order Blubervirales.

Mimivirus is a genus of giant viruses, in the family Mimiviridae. Amoeba serve as their natural hosts. This genus contains a single identified species named Acanthamoeba polyphaga mimivirus (APMV). It also refers to a group of phylogenetically related large viruses.
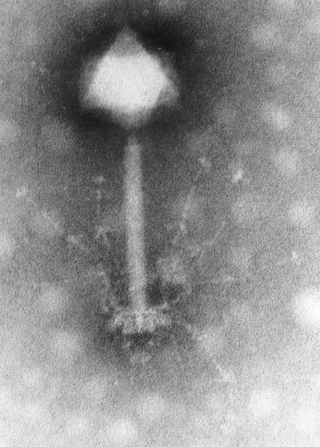
Myoviridae was a family of bacteriophages in the order Caudovirales. The family Myoviridae and order Caudovirales have now been abolished, with the term myovirus now used to refer to the morphology of viruses in this former family. Bacteria and archaea serve as natural hosts. There were 625 species in this family, assigned to eight subfamilies and 217 genera.

Filamentous bacteriophages are a family of viruses (Inoviridae) that infect bacteria, or bacteriophages. They are named for their filamentous shape, a worm-like chain, about 6 nm in diameter and about 1000-2000 nm long. This distinctive shape reflects their method of replication: the coat of the virion comprises five types of viral protein, which are located in the inner membrane of the host bacterium during phage assembly, and these proteins are added to the nascent virion's DNA as it is extruded through the membrane. The simplicity of filamentous phages makes them an appealing model organism for research in molecular biology, and they have also shown promise as tools in nanotechnology and immunology.

Geminiviridae is a family of plant viruses that encode their genetic information on a circular genome of single-stranded (ss) DNA. There are 520 species in this family, assigned to 14 genera. Diseases associated with this family include: bright yellow mosaic, yellow mosaic, yellow mottle, leaf curling, stunting, streaks, reduced yields. They have single-stranded circular DNA genomes encoding genes that diverge in both directions from a virion strand origin of replication. According to the Baltimore classification they are considered class II viruses. It is the largest known family of single stranded DNA viruses.

Parapoxvirus is a genus of viruses, in the family Poxviridae, in the subfamily Chordopoxvirinae. Like all members of the family Poxviridae, they are oval, relatively large, double-stranded DNA viruses. Parapoxviruses have a unique spiral coat that distinguishes them from other poxviruses. Parapoxviruses infect vertebrates, including a wide selection of mammals, and humans.

Mobile genetic elements (MGEs), sometimes called selfish genetic elements, are a type of genetic material that can move around within a genome, or that can be transferred from one species or replicon to another. MGEs are found in all organisms. In humans, approximately 50% of the genome is thought to be MGEs. MGEs play a distinct role in evolution. Gene duplication events can also happen through the mechanism of MGEs. MGEs can also cause mutations in protein coding regions, which alters the protein functions. These mechanisms can also rearrange genes in the host genome generating variation. These mechanism can increase fitness by gaining new or additional functions. An example of MGEs in evolutionary context are that virulence factors and antibiotic resistance genes of MGEs can be transported to share genetic code with neighboring bacteria. However, MGEs can also decrease fitness by introducing disease-causing alleles or mutations. The set of MGEs in an organism is called a mobilome, which is composed of a large number of plasmids, transposons and viruses.

Corticovirus is a genus of viruses in the family Corticoviridae. Corticoviruses are bacteriophages; that is, their natural hosts are bacteria. The genus contains two species. The name is derived from Latin cortex, corticis. However, prophages closely related to PM2 are abundant in the genomes of aquatic bacteria, suggesting that the ecological importance of corticoviruses might be underestimated. Bacteriophage PM2 was first described in 1968 after isolation from seawater sampled from the coast of Chile.

A virus is a submicroscopic infectious agent that replicates only inside the living cells of an organism. Viruses infect all life forms, from animals and plants to microorganisms, including bacteria and archaea. Viruses are found in almost every ecosystem on Earth and are the most numerous type of biological entity. Since Dmitri Ivanovsky's 1892 article describing a non-bacterial pathogen infecting tobacco plants and the discovery of the tobacco mosaic virus by Martinus Beijerinck in 1898, more than 11,000 of the millions of virus species have been described in detail. The study of viruses is known as virology, a subspeciality of microbiology.
Yatapoxvirus is a genus of viruses, in the family Poxviridae, in the subfamily Chordopoxvirinae. Monkeys and baboons serve as natural hosts. There are two species in this genus. Diseases associated with this genus include: histiocytomas, tumor-like mass of mononuclear cells.

Gene transfer agents (GTAs) are DNA-containing virus-like particles that are produced by some bacteria and archaea and mediate horizontal gene transfer. Different GTA types have originated independently from viruses in several bacterial and archaeal lineages. These cells produce GTA particles containing short segments of the DNA present in the cell. After the particles are released from the producer cell, they can attach to related cells and inject their DNA into the cytoplasm. The DNA can then become part of the recipient cells' genome.
Phikmvvirus is a genus of viruses that infect bacteria. There are currently 16 species in this genus including the type species Pseudomonas virus phiKMV. Bacteriophage phiKMV and its relatives are known to be highly virulent phages, producing large clear plaques on a susceptible host. The only reported exception is phage LKA1, which yields small plaques surrounded by a halo. While all other P. aeruginosa-specific phikmvviruses use the Type IV pili as primary receptor, LKA1 particles attach to the bacterial lipopolysaccharide layer.

Teseptimavirus is a genus of viruses in the order Caudovirales, in the family Autographiviridae, in the subfamily Studiervirinae. Bacteria serve as the natural host, with transmission achieved through passive diffusion. There are currently 17 species in this genus, including the type species Escherichia virus T7.
Spbetavirus is a genus of viruses in the order Caudovirales, in the family Siphoviridae. Bacteria serve as natural hosts. There is only one species in this genus: Bacillus virus SPbeta.
Cervidpoxvirus is a genus of viruses in the family Poxviridae in the subfamily Chordopoxvirinae. Deer serve as natural hosts. Only one species is in this genus: Mule deerpox virus.
Leporipoxvirus is a genus of viruses, in the family Poxviridae, in the subfamily Chordopoxvirinae. Lagomorphs and squirrels serve as natural hosts. There are four species in this genus. Diseases associated with this genus include: myxomatosis.
This glossary of virology is a list of definitions of terms and concepts used in virology, the study of viruses, particularly in the description of viruses and their actions. Related fields include microbiology, molecular biology, and genetics.

An archaeal virus is a virus that infects and replicates in archaea, a domain of unicellular, prokaryotic organisms. Archaeal viruses, like their hosts, are found worldwide, including in extreme environments inhospitable to most life such as acidic hot springs, highly saline bodies of water, and at the bottom of the ocean. They have been also found in the human body. The first known archaeal virus was described in 1974 and since then, a large diversity of archaeal viruses have been discovered, many possessing unique characteristics not found in other viruses. Little is known about their biological processes, such as how they replicate, but they are believed to have many independent origins, some of which likely predate the last archaeal common ancestor (LACA).












