In biology and biochemistry, protease inhibitors, or antiproteases, are molecules that inhibit the function of proteases. Many naturally occurring protease inhibitors are proteins.

Ku is a dimeric protein complex that binds to DNA double-strand break ends and is required for the non-homologous end joining (NHEJ) pathway of DNA repair. Ku is evolutionarily conserved from bacteria to humans. The ancestral bacterial Ku is a homodimer. Eukaryotic Ku is a heterodimer of two polypeptides, Ku70 (XRCC6) and Ku80 (XRCC5), so named because the molecular weight of the human Ku proteins is around 70 kDa and 80 kDa. The two Ku subunits form a basket-shaped structure that threads onto the DNA end. Once bound, Ku can slide down the DNA strand, allowing more Ku molecules to thread onto the end. In higher eukaryotes, Ku forms a complex with the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) to form the full DNA-dependent protein kinase, DNA-PK. Ku is thought to function as a molecular scaffold to which other proteins involved in NHEJ can bind, orienting the double-strand break for ligation.

A leucine-rich repeat (LRR) is a protein structural motif that forms an α/β horseshoe fold. It is composed of repeating 20–30 amino acid stretches that are unusually rich in the hydrophobic amino acid leucine. These tandem repeats commonly fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Typically, each repeat unit has beta strand-turn-alpha helix structure, and the assembled domain, composed of many such repeats, has a horseshoe shape with an interior parallel beta sheet and an exterior array of helices. One face of the beta sheet and one side of the helix array are exposed to solvent and are therefore dominated by hydrophilic residues. The region between the helices and sheets is the protein's hydrophobic core and is tightly sterically packed with leucine residues.

Carbamoyl phosphate synthetase catalyzes the ATP-dependent synthesis of carbamoyl phosphate from glutamine or ammonia and bicarbonate. This enzyme catalyzes the reaction of ATP and bicarbonate to produce carboxy phosphate and ADP. Carboxy phosphate reacts with ammonia to give carbamic acid. In turn, carbamic acid reacts with a second ATP to give carbamoyl phosphate plus ADP.
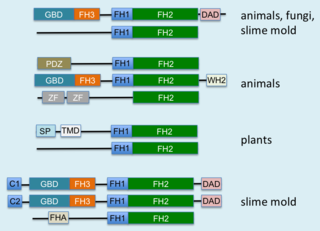
Formins are a group of proteins that are involved in the polymerization of actin and associate with the fast-growing end of actin filaments. Most formins are Rho-GTPase effector proteins. Formins regulate the actin and microtubule cytoskeleton and are involved in various cellular functions such as cell polarity, cytokinesis, cell migration and SRF transcriptional activity. Formins are multidomain proteins that interact with diverse signalling molecules and cytoskeletal proteins, although some formins have been assigned functions within the nucleus.

In the field of molecular biology, a two-component regulatory system serves as a basic stimulus-response coupling mechanism to allow organisms to sense and respond to changes in many different environmental conditions. Two-component systems typically consist of a membrane-bound histidine kinase that senses a specific environmental stimulus and a corresponding response regulator that mediates the cellular response, mostly through differential expression of target genes. Although two-component signaling systems are found in all domains of life, they are most common by far in bacteria, particularly in Gram-negative and cyanobacteria; both histidine kinases and response regulators are among the largest gene families in bacteria. They are much less common in archaea and eukaryotes; although they do appear in yeasts, filamentous fungi, and slime molds, and are common in plants, two-component systems have been described as "conspicuously absent" from animals.

In molecular biology, Phosphotyrosine-binding domains are protein domains which bind to phosphotyrosine.

The GHKL domain is an evolutionary conserved protein domain.
In molecular biology, the protein Sprouty is a developmental protein involved in cell signalling. It works by inhibiting the MAPK/ERK pathway.
In molecular biology the SPR domain is a protein domain found in the Sprouty (Spry) and Spread proteins. These have been identified as inhibitors of the Ras/mitogen-activated protein kinase (MAPK) cascade, a pathway crucial for developmental processes initiated by activation of various receptor tyrosine kinases. These proteins share a conserved, C-terminal cysteine-rich region, the SPR domain. This domain has been defined as a novel cytosol to membrane translocation domain. It has been found to be a PtdIns(4,5)P2-binding domain that targets the proteins to a cellular localization that maximizes their inhibitory potential. It also mediates homodimer formation of these proteins.
In molecular biology, autophagy related 3 (Atg3) is the E2 enzyme for the LC3 lipidation process. It is essential for autophagy. The super protein complex, the Atg16L complex, consists of multiple Atg12-Atg5 conjugates. Atg16L has an E3-like role in the LC3 lipidation reaction. The activated intermediate, LC3-Atg3 (E2), is recruited to the site where the lipidation takes place.
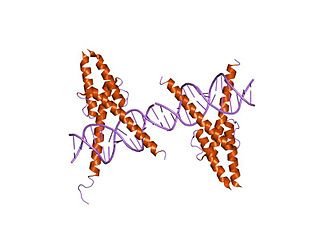
In molecular biology, the myogenic determination factor 5 proteins are a family of proteins found in eukaryotes. This family includes the Myf5 protein, which is responsible for directing cells to the skeletal myocyte lineage during development. Myf5 is likely to act in a similar way to the other MRF4 proteins such as MyoD which perform the same function. These are histone acetyltransferases and histone deacetylases which activate and repress genes involved in the myocyte lineage.

In molecular biology, the BPS domain domain is a protein domain of approximately 45 amino acids found in the adaptor proteins Grb7/|Grb10/Grb14. It mediates inhibition of the tyrosine kinase domain of the insulin receptor by binding of the N-terminal portion of the BPS domain to the substrate peptide groove of the kinase, acting as a pseudosubstrate inhibitor. It is composed of two beta strands and a C-terminal helix.

In molecular biology, the Btk-type zinc finger or Btk motif (BM) is a conserved zinc-binding motif containing conserved cysteines and a histidine that is present in certain eukaryotic signalling proteins. The motif is named after Bruton's tyrosine kinase (Btk), an enzyme which is essential for B cell maturation in humans and mice. Btk is a member of the Tec family of protein tyrosine kinases (PTK). These kinases contain a conserved Tec homology (TH) domain between the N-terminal pleckstrin homology (PH) domain and the Src homology 3 (SH3) domain. The N-terminal of the TH domain is highly conserved and known as the Btf motif, while the C-terminal region of the TH domain contains a proline-rich region (PRR). The Btk motif contains a conserved His and three Cys residues that form a zinc finger, while PRRs are commonly involved in protein-protein interactions, including interactions with G proteins. The TH domain may be of functional importance in various signalling pathways in different species. A complete TH domain, containing both the Btk and PRR regions, has not been found outside the Tec family; however, the Btk motif on its own does occur in other proteins, usually C-terminal to a PH domain.

In molecular biology, a carbohydrate-binding module (CBM) is a protein domain found in carbohydrate-active enzymes. The majority of these domains have carbohydrate-binding activity. Some of these domains are found on cellulosomal scaffoldin proteins. CBMs were previously known as cellulose-binding domains. CBMs are classified into numerous families, based on amino acid sequence similarity. There are currently 64 families of CBM in the CAZy database.
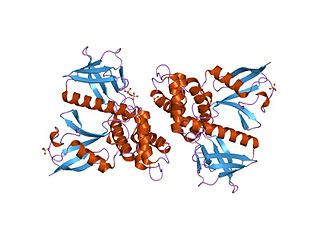
In molecular biology, the FERM domain is a widespread protein module involved in localising proteins to the plasma membrane. FERM domains are found in a number of cytoskeletal-associated proteins that associate with various proteins at the interface between the plasma membrane and the cytoskeleton. The FERM domain is located at the N terminus in the majority of proteins in which it is found.

In structural and cell biology, the focal adhesion targeting domain is a conserved protein domain that was first identified in focal adhesion kinase (FAK), also known as PTK2 protein tyrosine kinase 2 (PTK2).
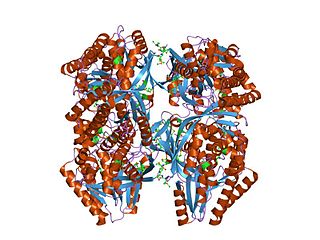
In molecular biology, the GHMP kinase family is a family of kinase enzymes. Members of this family include homoserine kinases EC 2.7.1.39, galactokinases EC 2.7.1.6, and mevalonate kinasesEC 2.7.1.36. These kinases make up the GHMP kinase superfamily of ATP-dependent enzymes. These enzymes are involved in the biosynthesis of isoprenes and amino acids as well as in carbohydrate metabolism. These enzymes contain, in their N-terminal section, a conserved Gly/Ser-rich region which is probably involved in the binding of ATP. The C-terminal domain of homoserine kinase has a central alpha-beta plait fold and an insertion of four helices, which, together with the N-terminal fold, creates a novel nucleotide binding fold.

In molecular biology, YopH, N-terminal refers to an evolutionary conserved protein domain. This entry represents the N-terminal domain of YopH protein tyrosine phosphatase (PTP).
In molecular biology, the protein domain, YTH refers to a member of the YTH family that has been shown to selectively remove transcripts of meiosis-specific genes expressed in mitotic cells.

















