Related Research Articles

Proteasomes are protein complexes which degrade unneeded or damaged proteins by proteolysis, a chemical reaction that breaks peptide bonds. Enzymes that help such reactions are called proteases.

Autophagy is the natural, conserved degradation of the cell that removes unnecessary or dysfunctional components through a lysosome-dependent regulated mechanism. It allows the orderly degradation and recycling of cellular components. Although initially characterized as a primordial degradation pathway induced to protect against starvation, it has become increasingly clear that autophagy also plays a major role in the homeostasis of non-starved cells. Defects in autophagy have been linked to various human diseases, including neurodegeneration and cancer, and interest in modulating autophagy as a potential treatment for these diseases has grown rapidly.

The bafilomycins are a family of macrolide antibiotics produced from a variety of Streptomycetes. Their chemical structure is defined by a 16-membered lactone ring scaffold. Bafilomycins exhibit a wide range of biological activity, including anti-tumor, anti-parasitic, immunosuppressant and anti-fungal activity. The most used bafilomycin is bafilomycin A1, a potent inhibitor of cellular autophagy. Bafilomycins have also been found to act as ionophores, transporting potassium K+ across biological membranes and leading to mitochondrial damage and cell death.
In eukaryotic cells, an aggresome refers to an aggregation of misfolded proteins in the cell, formed when the protein degradation system of the cell is overwhelmed. Aggresome formation is a highly regulated process that possibly serves to organize misfolded proteins into a single location.

Vojo Deretic, is distinguished professor and chair of the Department of Molecular Genetics and Microbiology at the University of New Mexico School of Medicine. Deretic was the founding director of the Autophagy, Inflammation and Metabolism (AIM) Center of Biomedical Research Excellence. The AIM center promotes autophagy research nationally and internationally.

Valosin-containing protein (VCP) or transitional endoplasmic reticulum ATPase also known as p97 in mammals and CDC48 in S. cerevisiae, is an enzyme that in humans is encoded by the VCP gene. The TER ATPase is an ATPase enzyme present in all eukaryotes and archaebacteria. Its main function is to segregate protein molecules from large cellular structures such as protein assemblies, organelle membranes and chromatin, and thus facilitate the degradation of released polypeptides by the multi-subunit protease proteasome.

STUB1 is a human gene that codes for the protein CHIP.

BAG family molecular chaperone regulator 3 is a protein that in humans is encoded by the BAG3 gene. BAG3 is involved in chaperone-assisted selective autophagy.

Autophagy protein 5 (ATG5) is a protein that, in humans, is encoded by the ATG5 gene located on chromosome 6. It is an E3 ubi autophagic cell death. ATG5 is a key protein involved in the extension of the phagophoric membrane in autophagic vesicles. It is activated by ATG7 and forms a complex with ATG12 and ATG16L1. This complex is necessary for LC3-I conjugation to PE (phosphatidylethanolamine) to form LC3-II. ATG5 can also act as a pro-apoptotic molecule targeted to the mitochondria. Under low levels of DNA damage, ATG5 can translocate to the nucleus and interact with survivin.

Microtubule-associated proteins 1A/1B light chain 3B is a protein that in humans is encoded by the MAP1LC3B gene. LC3 is a central protein in the autophagy pathway where it functions in autophagy substrate selection and autophagosome biogenesis. LC3 is the most widely used marker of autophagosomes.

Autophagy related 16 like 1 is a protein that in humans is encoded by the ATG16L1 gene. This protein is characterized as a subunit of the autophagy-related ATG12-ATG5/ATG16 complex and is essentially important for the LC3 (ATG8) lipidation and autophagosome formation. This complex localizes to the membrane and is released just before or after autophagosome completion.

In molecular biology, protein aggregation is a phenomenon in which intrinsically-disordered or mis-folded proteins aggregate either intra- or extracellularly. Protein aggregates have been implicated in a wide variety of diseases known as amyloidoses, including ALS, Alzheimer's, Parkinson's and prion disease.

Autophagy-related protein 8 (Atg8) is a ubiquitin-like protein required for the formation of autophagosomal membranes. The transient conjugation of Atg8 to the autophagosomal membrane through a ubiquitin-like conjugation system is essential for autophagy in eukaryotes. Even though there are homologues in animals, this article mainly focuses on its role in lower eukaryotes such as Saccharomyces cerevisiae.
Proteostasis is the dynamic regulation of a balanced, functional proteome. The proteostasis network includes competing and integrated biological pathways within cells that control the biogenesis, folding, trafficking, and degradation of proteins present within and outside the cell. Loss of proteostasis is central to understanding the cause of diseases associated with excessive protein misfolding and degradation leading to loss-of-function phenotypes, as well as aggregation-associated degenerative disorders. Therapeutic restoration of proteostasis may treat or resolve these pathologies.
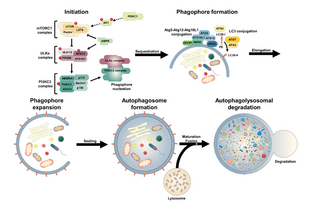
An autophagosome is a spherical structure with double layer membranes. It is the key structure in macroautophagy, the intracellular degradation system for cytoplasmic contents. After formation, autophagosomes deliver cytoplasmic components to the lysosomes. The outer membrane of an autophagosome fuses with a lysosome to form an autolysosome. The lysosome's hydrolases degrade the autophagosome-delivered contents and its inner membrane.
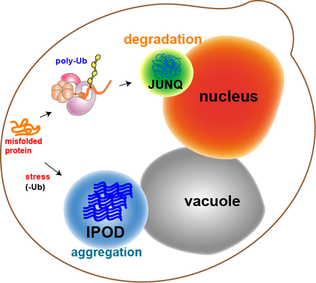
JUNQ and IPOD are types of cytosolic protein inclusion bodies in eukaryotes.

Chaperone-mediated autophagy (CMA) refers to the chaperone-dependent selection of soluble cytosolic proteins that are then targeted to lysosomes and directly translocated across the lysosome membrane for degradation. The unique features of this type of autophagy are the selectivity on the proteins that are degraded by this pathway and the direct shuttling of these proteins across the lysosomal membrane without the requirement for the formation of additional vesicles.

Omegasome is a cell organelle consisting of lipid bilayer membranes enriched for phosphatidylinositol 3-phosphate, and related to a process of autophagy. It is a subdomain of the Endoplasmic Reticulum (ER), and has a morphology resembling the Greek capital letter Omega (Ω). Omegasomes are the sites from which phagophores form, which are sack-like structures that mature into autophagosomes, and fuse with lysosomes in order to degrade the contents of the autophagosomes. The formation of omegasomes depends on various factors, however in general, formation of omegasomes is increased as a response to starvation, and in some biochemical situations the presence of PI(3)P leads to the formation of omegasomes.
Microautophagy is one of the three common forms of autophagic pathway, but unlike macroautophagy and chaperone-mediated autophagy, it is mediated—in mammals by lysosomal action or in plants and fungi by vacuolar action—by direct engulfment of the cytoplasmic cargo. Cytoplasmic material is trapped in the lysosome/vacuole by a random process of membrane invagination.
Atg8ylation is a process of conjugation of mammalian ATG8 proteins (mATG8s) to proteins or membranes. The process is akin to the ubiquitylation of diverse substrates by ubiquitin. There are six principal mATG8s: LC3A, LC3B, LC3C, GABARAP, GABARAPL1 and GABARAPL2. Together, they comprise a sub-class of ubiquitin-like molecules characterized by two N-terminal α-helices added to the ubiquitin core, which serve a dual role of forming a docking site for interacting proteins containing ATG8-interaction motifs and enhancing mATG8’s affinity for membranes.
References
- 1 2 3 4 5 Ulbricht A, Eppler FJ, Tapia VE, van der Ven PF, Hampe N, Hersch N, et al. (March 2013). "Cellular mechanotransduction relies on tension-induced and chaperone-assisted autophagy". Current Biology. 23 (5): 430–435. Bibcode:2013CBio...23..430U. doi: 10.1016/j.cub.2013.01.064 . PMID 23434281.
- 1 2 3 4 5 Arndt V, Dick N, Tawo R, Dreiseidler M, Wenzel D, Hesse M, et al. (January 2010). "Chaperone-assisted selective autophagy is essential for muscle maintenance". Current Biology. 20 (2): 143–148. Bibcode:2010CBio...20..143A. doi: 10.1016/j.cub.2009.11.022 . PMID 20060297.
- 1 2 3 Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, Behl C (April 2009). "Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3". The EMBO Journal. 28 (7): 889–901. doi:10.1038/emboj.2009.29. PMC 2647772 . PMID 19229298.
- 1 2 3 Carra S, Seguin SJ, Landry J (February 2008). "HspB8 and Bag3: a new chaperone complex targeting misfolded proteins to macroautophagy". Autophagy. 4 (2): 237–239. doi: 10.4161/auto.5407 . PMID 18094623.
- ↑ Reggiori F, Klionsky DJ (February 2002). "Autophagy in the eukaryotic cell". Eukaryotic Cell. 1 (1): 11–21. doi:10.1128/EC.01.1.11-21.2002. PMC 118053 . PMID 12455967.
- ↑ Levine B, Klionsky DJ (April 2004). "Development by self-digestion: molecular mechanisms and biological functions of autophagy". Developmental Cell. 6 (4): 463–477. doi: 10.1016/S1534-5807(04)00099-1 . PMID 15068787.
- 1 2 3 Shaid S, Brandts CH, Serve H, Dikic I (January 2013). "Ubiquitination and selective autophagy". Cell Death and Differentiation. 20 (1): 21–30. doi:10.1038/cdd.2012.72. PMC 3524631 . PMID 22722335.
- 1 2 3 4 Tedesco B, Vendredy L, Timmerman V, Poletti A (June 2023). "The chaperone-assisted selective autophagy complex dynamics and dysfunctions". Autophagy. 19 (6): 1619–1641. doi:10.1080/15548627.2022.2160564. PMC 10262806 . PMID 36594740.
- ↑ Selcen D, Muntoni F, Burton BK, Pegoraro E, Sewry C, Bite AV, Engel AG (January 2009). "Mutation in BAG3 causes severe dominant childhood muscular dystrophy". Annals of Neurology. 65 (1): 83–89. doi:10.1002/ana.21553. PMC 2639628 . PMID 19085932.
- ↑ Homma S, Iwasaki M, Shelton GD, Engvall E, Reed JC, Takayama S (September 2006). "BAG3 deficiency results in fulminant myopathy and early lethality". The American Journal of Pathology. 169 (3): 761–773. doi:10.2353/ajpath.2006.060250. PMC 1698816 . PMID 16936253.
- ↑ Sarparanta J, Jonson PH, Golzio C, Sandell S, Luque H, Screen M, et al. (February 2012). "Mutations affecting the cytoplasmic functions of the co-chaperone DNAJB6 cause limb-girdle muscular dystrophy". Nature Genetics. 44 (4): 450–5. doi:10.1038/ng.1103. PMC 3315599 . PMID 22366786.