Related Research Articles

An oncogene is a gene that has the potential to cause cancer. In tumor cells, these genes are often mutated, or expressed at high levels.

Cold Spring Harbor Laboratory (CSHL) is a private, non-profit institution with research programs focusing on cancer, neuroscience, plant biology, genomics, and quantitative biology. It is located in Laurel Hollow on Long Island, New York.

Acute myeloid leukemia (AML) is a cancer of the myeloid line of blood cells, characterized by the rapid growth of abnormal cells that build up in the bone marrow and blood and interfere with normal blood cell production. Symptoms may include feeling tired, shortness of breath, easy bruising and bleeding, and increased risk of infection. Occasionally, spread may occur to the brain, skin, or gums. As an acute leukemia, AML progresses rapidly, and is typically fatal within weeks or months if left untreated.
Chromatin remodeling is the dynamic modification of chromatin architecture to allow access of condensed genomic DNA to the regulatory transcription machinery proteins, and thereby control gene expression. Such remodeling is principally carried out by 1) covalent histone modifications by specific enzymes, e.g., histone acetyltransferases (HATs), deacetylases, methyltransferases, and kinases, and 2) ATP-dependent chromatin remodeling complexes which either move, eject or restructure nucleosomes. Besides actively regulating gene expression, dynamic remodeling of chromatin imparts an epigenetic regulatory role in several key biological processes, egg cells DNA replication and repair; apoptosis; chromosome segregation as well as development and pluripotency. Aberrations in chromatin remodeling proteins are found to be associated with human diseases, including cancer. Targeting chromatin remodeling pathways is currently evolving as a major therapeutic strategy in the treatment of several cancers.

Plicamycin is an antineoplastic antibiotic produced by Streptomyces plicatus. It is an RNA synthesis inhibitor. The manufacturer discontinued production in 2000. Several different structures are currently reported in different places all with the same chromomycin core, but with different stereochemistry in the glycoside chain, a 1999 study has re-investigated the compound and proposed a revised structure.
Runt-related transcription factor 1 (RUNX1) also known as acute myeloid leukemia 1 protein (AML1) or core-binding factor subunit alpha-2 (CBFA2) is a protein that in humans is encoded by the RUNX1 gene.

CCAAT/enhancer-binding protein alpha is a protein encoded by the CEBPA gene in humans. CCAAT/enhancer-binding protein alpha is a transcription factor involved in the differentiation of certain blood cells. For details on the CCAAT structural motif in gene enhancers and on CCAAT/Enhancer Binding Proteins see the specific page.

CD47 also known as integrin associated protein (IAP) is a transmembrane protein that in humans is encoded by the CD47 gene. CD47 belongs to the immunoglobulin superfamily and partners with membrane integrins and also binds the ligands thrombospondin-1 (TSP-1) and signal-regulatory protein alpha (SIRPα). CD-47 acts as a don't eat me signal to macrophages of the immune system which has made it a potential therapeutic target in some cancers, and more recently, for the treatment of pulmonary fibrosis.

Sal-like protein 4(SALL4) is a transcription factor encoded by a member of the Spalt-like (SALL) gene family, SALL4. The SALL genes were identified based on their sequence homology to Spalt, which is a homeotic gene originally cloned in Drosophila melanogaster that is important for terminal trunk structure formation in embryogenesis and imaginal disc development in the larval stages. There are four human SALL proteins with structural homology and playing diverse roles in embryonic development, kidney function, and cancer. The SALL4 gene encodes at least three isoforms, termed A, B, and C, through alternative splicing, with the A and B forms being the most studied. SALL4 can alter gene expression changes through its interaction with many co-factors and epigenetic complexes. It is also known as a key embryonic stem cell (ESC) factor.

Bromodomain-containing protein 4 is a protein that in humans is encoded by the BRD4 gene.

Cancer epigenetics is the study of epigenetic modifications to the DNA of cancer cells that do not involve a change in the nucleotide sequence, but instead involve a change in the way the genetic code is expressed. Epigenetic mechanisms are necessary to maintain normal sequences of tissue specific gene expression and are crucial for normal development. They may be just as important, if not even more important, than genetic mutations in a cell's transformation to cancer. The disturbance of epigenetic processes in cancers, can lead to a loss of expression of genes that occurs about 10 times more frequently by transcription silencing than by mutations. As Vogelstein et al. points out, in a colorectal cancer there are usually about 3 to 6 driver mutations and 33 to 66 hitchhiker or passenger mutations. However, in colon tumors compared to adjacent normal-appearing colonic mucosa, there are about 600 to 800 heavily methylated CpG islands in the promoters of genes in the tumors while these CpG islands are not methylated in the adjacent mucosa. Manipulation of epigenetic alterations holds great promise for cancer prevention, detection, and therapy. In different types of cancer, a variety of epigenetic mechanisms can be perturbed, such as the silencing of tumor suppressor genes and activation of oncogenes by altered CpG island methylation patterns, histone modifications, and dysregulation of DNA binding proteins. There are several medications which have epigenetic impact, that are now used in a number of these diseases.

JQ1 is a thienotriazolodiazepine and a potent inhibitor of the BET family of bromodomain proteins which include BRD2, BRD3, BRD4, and the testis-specific protein BRDT in mammals. BET inhibitors structurally similar to JQ1 are being tested in clinical trials for a variety of cancers including NUT midline carcinoma. It was developed by the James Bradner laboratory at Brigham and Women's Hospital and named after chemist Jun Qi. The chemical structure was inspired by patent of similar BET inhibitors by Mitsubishi Tanabe Pharma. Structurally it is related to benzodiazepines. While widely used in laboratory applications, JQ1 is not itself being used in human clinical trials because it has a short half life.
BET inhibitors are a class of drugs that reversibly bind the bromodomains of Bromodomain and Extra-Terminal motif (BET) proteins BRD2, BRD3, BRD4, and BRDT, and prevent protein-protein interaction between BET proteins and acetylated histones and transcription factors.
David Arthur Tuveson is an American cancer biologist and is currently Roy J. Zuckerberg Professor of Cancer Research as well as The Cancer Center Director at Cold Spring Harbor Laboratory. Dr. Tuveson is also the Chief Scientist for the Lustgarten Foundation for Pancreatic Cancer Research. He is known for developing some of the first mouse models of pancreatic cancer and more recently, for his work developing pancreatic cancer organoids.
Prognostic markers are biomarkers used to measure the progress of a disease in the patient sample. Prognostic markers are useful to stratify the patients into groups, guiding towards precise medicine discovery. The widely used prognostic markers in cancers include stage, size, grade, node and metastasis. In addition to these common markers, there are prognostic markers specific to different cancer types. For example estrogen level, progesterone and HER2 are markers specific to breast cancer patients. There is evidence showing that genes behaving as tumor suppressors or carcinogens could act as prognostic markers due to altered gene expression or mutation. Besides genetic biomarkers, there are also biomarkers that are detected in plasma or body fluid which can be metabolic or protein biomarkers.
Dafna Bar-Sagi is a cell biologist and cancer researcher at New York University School of Medicine. She is the Saul J. Farber Professor in the department of biochemistry and molecular pharmacology and the department of medicine and senior vice president and vice dean for science at NYU Langone Health. Bar-Sagi has been a member of scientific advisory boards, including the National Cancer Institute, Starr Cancer Consortium, and Pancreatic Cancer Action Network.
Jason Sheltzer is a cancer biologist at the Yale University School of Medicine.
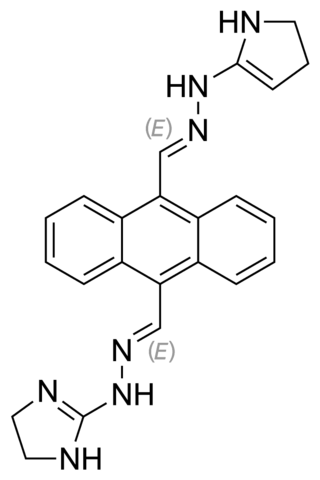
Bisantrene is an anthracenyl bishydrazone with anthracycline-like antineoplastic activity and an antimetabolite. Bisantrene intercalates with and disrupts the configuration of DNA, resulting in DNA single-strand breaks, DNA-protein crosslinking, and inhibition of DNA replication. This agent is similar to doxorubicin in chemotherapeutic activity, but unlike anthracyclines like doxorubicin, it exhibits little cardiotoxicity.

Oncometabolism is the field of study that focuses on the metabolic changes that occur in cells that make up the tumor microenvironment (TME) and accompany oncogenesis and tumor progression toward a neoplastic state.
The nuclear protein in testis gene encodes a 1,132-amino acid protein termed NUT that is expressed almost exclusively in the testes, ovaries, and ciliary ganglion. NUT protein facilitates the acetylation of chromatin by histone acetyltransferase EP300 in testicular spermatids. This acetylation is a form of chromatin remodeling which compacts spermatid chromatin, a critical step required for the normal conduct of spermatogenesis, i.e. the maturation of spermatids into sperm. Male mice that lacked the mouse Nutm1 gene using a gene knockout method had abnormally small testes, lacked sperm in their cauda epididymis, and were completely sterile. These findings indicate that Nutm1 gene is essential for the development of normal fertility in male mice and suggest that the NUTM1 gene may play a similar role in men.
References
- 1 2 3 4 5 6 "Christopher Vakoc, MD, PhD". Pershing Square Foundation. Retrieved 2020-05-01.
- 1 2 Ryan, Joe (2013-06-07). "Cold Spring Harbor Lab targeting cancer cells". Newsday. Archived from the original on 2013-09-01. Retrieved 2020-05-04.
- ↑ "A Conversation with Christopher Vakoc". Cold Spring Harbor Symposia on Quantitative Biology. 81: 344–346. 2016-01-01. doi: 10.1101/sqb.2016.81.031617 . ISSN 0091-7451. PMID 28123048.
- ↑ Zuber, Johannes; Shi, Junwei; Wang, Eric; Rappaport, Amy R.; Herrmann, Harald; Sison, Edward A.; Magoon, Daniel; Qi, Jun; Blatt, Katharina; Wunderlich, Mark; Taylor, Meredith J. (October 2011). "RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia". Nature. 478 (7370): 524–528. Bibcode:2011Natur.478..524Z. doi:10.1038/nature10334. ISSN 1476-4687. PMC 3328300 . PMID 21814200.
- ↑ Ricks, Delthia (2015-04-18). "Cold Spring Harbor researchers test treatment that can halt acute myeloid leukemia". Newsday. Retrieved 2020-05-01.
- ↑ Ricks, Delthia (2018-07-14). "Cold Spring Harbor scientists discover new form of lung cancer". Newsday. Retrieved 2020-05-01.
- ↑ Kwon, Diana (2017-07-27). "Enhancers Drive Pancreatic Cancer Metastasis: Study". The Scientist . Retrieved 2020-05-01.
- ↑ "LI scientists make key pancreatic cancer find". Newsday. Retrieved 2020-05-04.
- ↑ Rood, Jenny (2015-05-13). "Targeting Protein Domains with CRISPR". The Scientist. Retrieved 2020-05-01.
- ↑ Howley, Elaine K. (2018-09-12). "What's the likelihood that CRISPR will cure Cancer?". U.S. News & World Report. Archived from the original on 2018-09-13.
- ↑ "AACR recognizes outstanding cancer research achievements of Dr. Christopher Vakoc". EurekAlert!. Retrieved 2020-05-04.
- ↑ Grisham, Julie (2019-11-08). "Three Scientists are Named Winners of the Paul Marks Prize for Cancer Research". Memorial Sloan Kettering Cancer Center. Retrieved 2020-05-03.