Related Research Articles
An allele, or allelomorph, is a variant of the sequence of nucleotides at a particular location, or locus, on a DNA molecule.

An autosome is any chromosome that is not a sex chromosome. The members of an autosome pair in a diploid cell have the same morphology, unlike those in allosomal pairs, which may have different structures. The DNA in autosomes is collectively known as atDNA or auDNA.
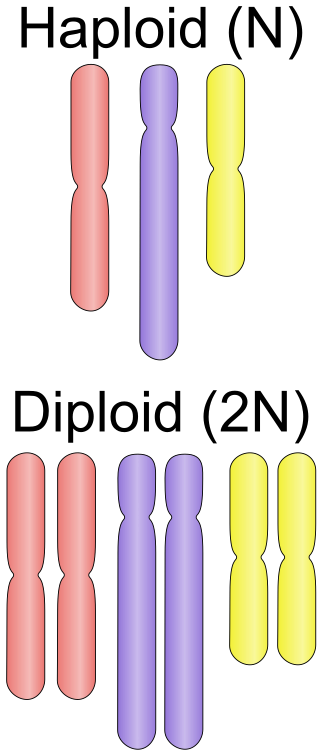
Ploidy is the number of complete sets of chromosomes in a cell, and hence the number of possible alleles for autosomal and pseudoautosomal genes. Sets of chromosomes refer to the number of maternal and paternal chromosome copies, respectively, in each homologous chromosome pair, which chromosomes naturally exist as. Somatic cells, tissues, and individual organisms can be described according to the number of sets of chromosomes present : monoploid, diploid, triploid, tetraploid, pentaploid, hexaploid, heptaploid or septaploid, etc. The generic term polyploid is often used to describe cells with three or more sets of chromosomes.
In molecular biology, restriction fragment length polymorphism (RFLP) is a technique that exploits variations in homologous DNA sequences, known as polymorphisms, populations, or species or to pinpoint the locations of genes within a sequence. The term may refer to a polymorphism itself, as detected through the differing locations of restriction enzyme sites, or to a related laboratory technique by which such differences can be illustrated. In RFLP analysis, a DNA sample is digested into fragments by one or more restriction enzymes, and the resulting restriction fragments are then separated by gel electrophoresis according to their size.

A karyotype is the general appearance of the complete set of chromosomes in the cells of a species or in an individual organism, mainly including their sizes, numbers, and shapes. Karyotyping is the process by which a karyotype is discerned by determining the chromosome complement of an individual, including the number of chromosomes and any abnormalities.

Cytogenetics is essentially a branch of genetics, but is also a part of cell biology/cytology, that is concerned with how the chromosomes relate to cell behaviour, particularly to their behaviour during mitosis and meiosis. Techniques used include karyotyping, analysis of G-banded chromosomes, other cytogenetic banding techniques, as well as molecular cytogenetics such as fluorescence in situ hybridization (FISH) and comparative genomic hybridization (CGH).

In biology, polymorphism is the occurrence of two or more clearly different morphs or forms, also referred to as alternative phenotypes, in the population of a species. To be classified as such, morphs must occupy the same habitat at the same time and belong to a panmictic population.

In genetics, chromosome translocation is a phenomenon that results in unusual rearrangement of chromosomes. This includes balanced and unbalanced translocation, with two main types: reciprocal, and Robertsonian translocation. Reciprocal translocation is a chromosome abnormality caused by exchange of parts between non-homologous chromosomes. Two detached fragments of two different chromosomes are switched. Robertsonian translocation occurs when two non-homologous chromosomes get attached, meaning that given two healthy pairs of chromosomes, one of each pair "sticks" and blends together homogeneously.

A haplotype is a group of alleles in an organism that are inherited together from a single parent.
Balancing selection refers to a number of selective processes by which multiple alleles are actively maintained in the gene pool of a population at frequencies larger than expected from genetic drift alone. Balancing selection is rare compared to purifying selection. It can occur by various mechanisms, in particular, when the heterozygotes for the alleles under consideration have a higher fitness than the homozygote. In this way genetic polymorphism is conserved.

Robertsonian translocation (ROB) is a chromosomal abnormality where the entire long arms of two different chromosomes become fused to each other. It is the most common form of chromosomal translocation in humans, affecting 1 out of every 1,000 babies born. It does not usually cause medical problems, though some people may produce gametes with an incorrect number of chromosomes, resulting in a risk of miscarriage. In rare cases this translocation results in Down syndrome and Patau syndrome. Robertsonian translocations result in a reduction in the number of chromosomes. A Robertsonian evolutionary fusion, which may have occurred in the common ancestor of humans and other great apes, is the reason humans have 46 chromosomes while all other primates have 48. Detailed DNA studies of chimpanzee, orangutan, gorilla and bonobo apes has determined that where human chromosome 2 is present in our DNA in all four great apes this is split into two separate chromosomes typically numbered 2a and 2b. Similarly, the fact that horses have 64 chromosomes and donkeys 62, and that they can still have common, albeit usually infertile, offspring, may be due to a Robertsonian evolutionary fusion at some point in the descent of today's donkeys from their common ancestor.

An inversion is a chromosome rearrangement in which a segment of a chromosome becomes inverted within its original position. An inversion occurs when a chromosome undergoes a two breaks within the chromosomal arm, and the segment between the two breaks inserts itself in the opposite direction in the same chromosome arm. The breakpoints of inversions often happen in regions of repetitive nucleotides, and the regions may be reused in other inversions. Chromosomal segments in inversions can be as small as 100 kilobases or as large as 100 megabases. The number of genes captured by an inversion can range from a handful of genes to hundreds of genes. Inversions can happen either through ectopic recombination, chromosomal breakage and repair, or non-homologous end joining.
Human genetics is the study of inheritance as it occurs in human beings. Human genetics encompasses a variety of overlapping fields including: classical genetics, cytogenetics, molecular genetics, biochemical genetics, genomics, population genetics, developmental genetics, clinical genetics, and genetic counseling.
Genetic variability is either the presence of, or the generation of, genetic differences. It is defined as "the formation of individuals differing in genotype, or the presence of genotypically different individuals, in contrast to environmentally induced differences which, as a rule, cause only temporary, nonheritable changes of the phenotype". Genetic variability in a population is important for biodiversity.
The mechanisms of reproductive isolation are a collection of evolutionary mechanisms, behaviors and physiological processes critical for speciation. They prevent members of different species from producing offspring, or ensure that any offspring are sterile. These barriers maintain the integrity of a species by reducing gene flow between related species.
A dicentric chromosome is an abnormal chromosome with two centromeres. It is formed through the fusion of two chromosome segments, each with a centromere, resulting in the loss of acentric fragments and the formation of dicentric fragments. The formation of dicentric chromosomes has been attributed to genetic processes, such as Robertsonian translocation and paracentric inversion. Dicentric chromosomes have important roles in the mitotic stability of chromosomes and the formation of pseudodicentric chromosomes. Their existence has been linked to certain natural phenomena such as irradiation and have been documented to underlie certain clinical syndromes, notably Kabuki syndrome. The formation of dicentric chromosomes and their implications on centromere function are studied in certain clinical cytogenetics laboratories.

Hybrid speciation is a form of speciation where hybridization between two different species leads to a new species, reproductively isolated from the parent species. Previously, reproductive isolation between two species and their parents was thought to be particularly difficult to achieve, and thus hybrid species were thought to be very rare. With DNA analysis becoming more accessible in the 1990s, hybrid speciation has been shown to be a somewhat common phenomenon, particularly in plants. In botanical nomenclature, a hybrid species is also called a nothospecies. Hybrid species are by their nature polyphyletic.
In genetics, pseudolinkage is a characteristic of a heterozygote for a reciprocal translocation, in which genes located near the translocation breakpoint behave as if they are linked even though they originated on nonhomologous chromosomes.
The Infinite sites model (ISM) is a mathematical model of molecular evolution first proposed by Motoo Kimura in 1969. Like other mutation models, the ISM provides a basis for understanding how mutation develops new alleles in DNA sequences. Using allele frequencies, it allows for the calculation of heterozygosity, or genetic diversity, in a finite population and for the estimation of genetic distances between populations of interest.
This glossary of genetics and evolutionary biology is a list of definitions of terms and concepts used in the study of genetics and evolutionary biology, as well as sub-disciplines and related fields, with an emphasis on classical genetics, quantitative genetics, population biology, phylogenetics, speciation, and systematics. Overlapping and related terms can be found in Glossary of cellular and molecular biology, Glossary of ecology, and Glossary of biology.
References
- ↑ Borin, Luciana Andreia; Isabel Cristina Martins-Santos (September 2000). "Intra-individual numerical chromosomal polymorphism in Trichomycterus davisi (Siluriformes, Trichomycteridae) from the Iguaçu River basin in Brazil" (PDF). Genetics and Molecular Biology. 23 (3): 605–607. doi: 10.1590/S1415-47572000000300018 .
- ↑ Bauchan, Gary R.; T. Austin Campbell; M. Azhar Hossain (July 1, 2002). "Chromosomal Polymorphism as Detected by C-Banding Patterns in Chilean Alfalfa Germplasm". Crop Science. 42 (4): 1291–7. doi:10.2135/cropsci2002.1291. S2CID 85405717. Archived from the original on December 1, 2005. Retrieved November 10, 2005.
- ↑ Elrod DA, Beck ML, Kennedy ML (October 1996). "Chromosomal variation in the southern short-tailed shrew (Blarina carolinensis)". Genetica. 98 (2): 199–203. doi:10.1007/BF00121367. PMID 8999000. S2CID 20111391.
- ↑ Thales Renato O. de Freitas (1997). "Chromosome polymorphism in Ctenomys minutus (Rodentia-Octodontidae)". Brazilian Journal of Genetics. 20 (1). doi: 10.1590/S0100-84551997000100001 .
- ↑ "Google Scholar".
- ↑ Barry Starr (February 26, 2010). "The 44 Chromosome Man And What He Reveals About Our Genetic Past". The Tech Museum. Archived from the original on December 6, 2011.