
In molecular biology, U3 snoRNA is a non-coding RNA found predominantly in the nucleolus. U3 has C/D box motifs that technically make it a member of the box C/D class of snoRNAs; however, unlike other C/D box snoRNAs, it has not been shown to direct 2'-O-methylation of other RNAs. Rather, U3 is thought to guide site-specific cleavage of ribosomal RNA (rRNA) during pre-rRNA processing.

The U4 small nuclear Ribo-Nucleic Acid is a non-coding RNA component of the major or U2-dependent spliceosome – a eukaryotic molecular machine involved in the splicing of pre-messenger RNA (pre-mRNA). It forms a duplex with U6, and with each splicing round, it is displaced from the U6 snRNA in an ATP-dependent manner, allowing U6 to re-fold and create the active site for splicing catalysis. A recycling process involving protein Brr2 releases U4 from U6, while protein Prp24 re-anneals U4 and U6. The crystal structure of a 5′ stem-loop of U4 in complex with a binding protein has been solved.

In molecular biology, heat shock factors (HSF), are the transcription factors that regulate the expression of the heat shock proteins. A typical example is the heat shock factor of Drosophila melanogaster.
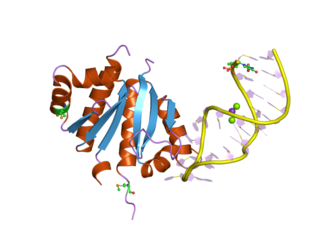
The K Homology (KH) domain is a protein domain that was first identified in the human heterogeneous nuclear ribonucleoprotein (hnRNP) K. An evolutionarily conserved sequence of around 70 amino acids, the KH domain is present in a wide variety of nucleic acid-binding proteins. The KH domain binds RNA, and can function in RNA recognition. It is found in multiple copies in several proteins, where they can function cooperatively or independently. For example, in the AU-rich element RNA-binding protein KSRP, which has 4 KH domains, KH domains 3 and 4 behave as independent binding modules to interact with different regions of the AU-rich RNA targets. The solution structure of the first KH domain of FMR1 and of the C-terminal KH domain of hnRNP K determined by nuclear magnetic resonance (NMR) revealed a beta-alpha-alpha-beta-beta-alpha structure. Autoantibodies to NOVA1, a KH domain protein, cause paraneoplastic opsoclonus ataxia. The KH domain is found at the N-terminus of the ribosomal protein S3. This domain is unusual in that it has a different fold compared to the normal KH domain.

Heterogeneous nuclear ribonucleoprotein A1 is a protein that in humans is encoded by the HNRNPA1 gene. Mutations in hnRNP A1 are causative of amyotrophic lateral sclerosis and the syndrome multisystem proteinopathy.

Poly(rC)-binding protein 1 is a protein that in humans is encoded by the PCBP1 gene.

Poly(rC)-binding protein 2 is a protein that in humans is encoded by the PCBP2 gene.

DEAD box proteins are involved in an assortment of metabolic processes that typically involve RNAs, but in some cases also other nucleic acids. They are highly conserved in nine motifs and can be found in most prokaryotes and eukaryotes, but not all. Many organisms, including humans, contain DEAD-box (SF2) helicases, which are involved in RNA metabolism.
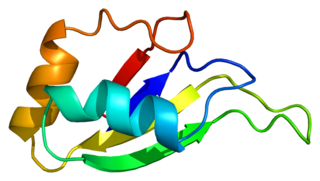
Heterogeneous nuclear ribonucleoprotein D0 (HNRNPD) also known as AU-rich element RNA-binding protein 1 (AUF1) is a protein that in humans is encoded by the HNRNPD gene. Alternative splicing of this gene results in four transcript variants.

Heterogeneous nuclear ribonucleoproteins C1/C2 is a protein that in humans is encoded by the HNRNPC gene.

Small nuclear ribonucleoprotein-associated proteins B and B' is a protein that in humans is encoded by the SNRPB gene.

Splicing factor 3 subunit 1 is a protein that in humans is encoded by the SF3A1 gene.
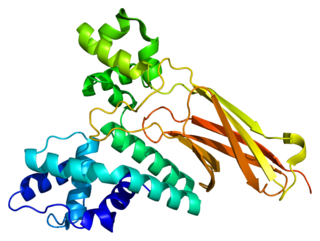
U5 small nuclear ribonucleoprotein 200 kDa helicase is an enzyme that in humans is encoded by the SNRNP200 gene.

H/ACA ribonucleoprotein complex subunit 2 is a protein that in humans is encoded by the NHP2 gene.

Cold shock domain-containing protein C2 is a protein that in humans is encoded by the CSDC2 gene.

Prp24 is a protein part of the pre-messenger RNA splicing process and aids the binding of U6 snRNA to U4 snRNA during the formation of spliceosomes. Found in eukaryotes from yeast to E. coli, fungi, and humans, Prp24 was initially discovered to be an important element of RNA splicing in 1989. Mutations in Prp24 were later discovered in 1991 to suppress mutations in U4 that resulted in cold-sensitive strains of yeast, indicating its involvement in the reformation of the U4/U6 duplex after the catalytic steps of splicing.

RNA recognition motif, RNP-1 is a putative RNA-binding domain of about 90 amino acids that are known to bind single-stranded RNAs. It was found in many eukaryotic proteins.
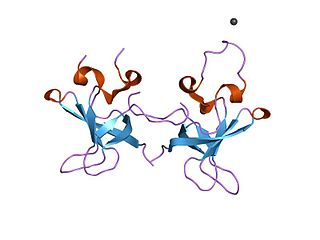
The S1 domain is a protein domain that was originally identified in ribosomal protein S1 but is found in a large number of RNA-associated proteins. The structure of the S1 RNA-binding domain from the Escherichia coli polynucleotide phosphorylase has been determined using NMR methods and consists of a five-stranded antiparallel beta barrel. Conserved residues on one face of the barrel and adjacent loops form the putative RNA-binding site.
The RNA-binding Proteins Database (RBPDB) is a biological database of RNA-binding protein specificities that includes experimental observations of RNA-binding sites. The experimental results included are both in vitro and in vivo from primary literature. It includes four metazoan species, which are Homo sapiens, Mus musculus, Drosophila melanogaster, and Caenorhabditis elegans. RNA-binding domains included in this database are RNA recognition motif, K homology, CCCH zinc finger, and more domains. As of 2021, the latest RBPDB release includes 1,171 RNA-binding proteins.
The arginine-glycine or arginine-glycine-glycine (RG/RGG) motif is a repeating amino acid sequence motif commonly found in RNA-binding proteins (RBPs). RGG regions in proteins are defined as two or more RG/RGG sequences within a stretch of 30 amino acids. Initially named the RGG box, it confers a protein with the ability to bind double-stranded mRNA molecules. The RGG motif has been observed in proteins from at least 12 animal species, including humans.


















