This article may be too technical for most readers to understand.(May 2015) |
Cospin is a serine protease inhibitor from the mushroom species Coprinopsis cinerea in the phylum Basidiomycota.
This article may be too technical for most readers to understand.(May 2015) |
Cospin is a serine protease inhibitor from the mushroom species Coprinopsis cinerea in the phylum Basidiomycota.
It has similar biochemical properties to other well characterized fungal serine protease inhibitors of family I66 in the MEROPS classification. [1] [2] [3] [4] Cospin is one of the few serine protease inhibitors of basidiomycete that have been isolated and characterized. [5] It is highly expressed in sexual reproductive structures termed fruiting bodies and has been cloned and characterized at the molecular and functional levels.
The cospin PIC 1 gene sequence isolated from fruiting bodies of the C. cinerea strain AmutBmut can be found under GenBank TM accession number ACX48485. [6]
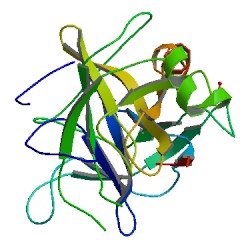
Cospin is a small protein and a highly specific trypsin inhibitor. pH stability Recombinant purified Cospin has been found to remain active after incubation in acidic pH 3 and alkaline pH 11 conditions. [6] Crystal structure Cospin is based on a β-trefoil fold. [6] The cospin fold resembles a tree-like structure with 2 loops in the root region, a stem comprising a six-stranded β-barrel, and two layers of loops in the crown region. [6] Cospin, is the first fungal trypsin inhibitor with a determined 3D structure, it utilizes a different loop for trypsin inhibition compared to other beta-trefoil inhibitors. [7] Substrate binding“cospin is a classic inhibitor that binds to the active site in a substrate-like manner and forms a tight and stable complex with trypsin.” [6] Cospin biotoxicity is not lethal to amoeba A.castellanii, nematode Caenorhabditis elegans, and two true fly species. The toxicity caused developmental delay in both pupae and flies. [6]
Cospin plays a defensive role against predators and parasites. [6] Cospin toxicity targets specifically fruit fly D. melanogaster. [7] Cospin represents one type of fungal protein mediated defence against fungivorous insects. [6] Cospin a serine protease inhibitor can be of use in various applications ranging from pest management, crop protection to drug development, design for therapeutics, and basic medical research. [6]
Chymotrypsin (EC 3.4.21.1, chymotrypsins A and B, alpha-chymar ophth, avazyme, chymar, chymotest, enzeon, quimar, quimotrase, alpha-chymar, alpha-chymotrypsin A, alpha-chymotrypsin) is a digestive enzyme component of pancreatic juice acting in the duodenum, where it performs proteolysis, the breakdown of proteins and polypeptides. Chymotrypsin preferentially cleaves peptide amide bonds where the side chain of the amino acid N-terminal to the scissile amide bond (the P1 position) is a large hydrophobic amino acid (tyrosine, tryptophan, and phenylalanine). These amino acids contain an aromatic ring in their side chain that fits into a hydrophobic pocket (the S1 position) of the enzyme. It is activated in the presence of trypsin. The hydrophobic and shape complementarity between the peptide substrate P1 side chain and the enzyme S1 binding cavity accounts for the substrate specificity of this enzyme. Chymotrypsin also hydrolyzes other amide bonds in peptides at slower rates, particularly those containing leucine and methionine at the P1 position.

Trypsin is an enzyme in the first section of the small intestine that starts the digestion of protein molecules by cutting these long chains of amino acids into smaller pieces. It is a serine protease from the PA clan superfamily, found in the digestive system of many vertebrates, where it hydrolyzes proteins. Trypsin is formed in the small intestine when its proenzyme form, the trypsinogen produced by the pancreas, is activated. Trypsin cuts peptide chains mainly at the carboxyl side of the amino acids lysine or arginine. It is used for numerous biotechnological processes. The process is commonly referred to as trypsin proteolysis or trypsinization, and proteins that have been digested/treated with trypsin are said to have been trypsinized. Trypsin was discovered in 1876 by Wilhelm Kühne and was named from the Ancient Greek word for rubbing since it was first isolated by rubbing the pancreas with glycerin.

A protease is an enzyme that catalyzes proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks bonds. Proteases are involved in many biological functions, including digestion of ingested proteins, protein catabolism, and cell signaling.
In biology and biochemistry, protease inhibitors, or antiproteases, are molecules that inhibit the function of proteases. Many naturally occurring protease inhibitors are proteins.

Serine proteases are enzymes that cleave peptide bonds in proteins. Serine serves as the nucleophilic amino acid at the (enzyme's) active site. They are found ubiquitously in both eukaryotes and prokaryotes. Serine proteases fall into two broad categories based on their structure: chymotrypsin-like (trypsin-like) or subtilisin-like.

A catalytic triad is a set of three coordinated amino acids that can be found in the active site of some enzymes. Catalytic triads are most commonly found in hydrolase and transferase enzymes. An acid-base-nucleophile triad is a common motif for generating a nucleophilic residue for covalent catalysis. The residues form a charge-relay network to polarise and activate the nucleophile, which attacks the substrate, forming a covalent intermediate which is then hydrolysed to release the product and regenerate free enzyme. The nucleophile is most commonly a serine or cysteine amino acid, but occasionally threonine or even selenocysteine. The 3D structure of the enzyme brings together the triad residues in a precise orientation, even though they may be far apart in the sequence.

Cysteine proteases, also known as thiol proteases, are hydrolase enzymes that degrade proteins. These proteases share a common catalytic mechanism that involves a nucleophilic cysteine thiol in a catalytic triad or dyad.

Maspin is a protein that in humans is encoded by the SERPINB5 gene. This protein belongs to the serpin superfamily. SERPINB5 was originally reported to function as a tumor suppressor gene in epithelial cells, suppressing the ability of cancer cells to invade and metastasize to other tissues. Furthermore, and consistent with an important biological function, Maspin knockout mice were reported to be non-viable, dying in early embryogenesis. However, a subsequent study using viral transduction as a method of gene transfer was not able to reproduce the original findings and found no role for maspin in tumour biology. Furthermore, the latter study demonstrated that maspin knockout mice are viable and display no obvious phenotype. These data are consistent with the observation that maspin is not expressed in early embryogenesis. The precise molecular function of maspin is thus currently unknown.

Serine protease HTRA2, mitochondrial is an enzyme that in humans is encoded by the HTRA2 gene. This protein is involved in caspase-dependent apoptosis and in Parkinson's disease.
Lympho-epithelial Kazal-type-related inhibitor (LEKTI) also known as serine protease inhibitor Kazal-type 5 (SPINK5) is a protein that in humans is encoded by the SPINK5 gene.

Kallikrein-14 is a protein that in humans is encoded by the KLK14 gene.
The Proteolysis MAP (PMAP) is an integrated web resource focused on proteases.
Kunitz soybean trypsin inhibitor is a type of protein contained in legume seeds which functions as a protease inhibitor. Kunitz-type Soybean Trypsin Inhibitors are usually specific for either trypsin or chymotrypsin. They are thought to protect seeds against consumption by animal predators.
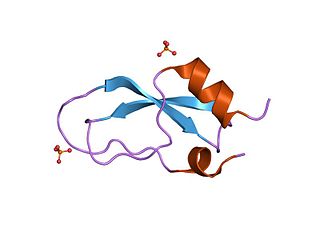
Kunitz domains are the active domains of proteins that inhibit the function of protein degrading enzymes or, more specifically, domains of Kunitz-type are protease inhibitors. They are relatively small with a length of about 50 to 60 amino acids and a molecular weight of 6 kDa. Examples of Kunitz-type protease inhibitors are aprotinin, Alzheimer's amyloid precursor protein (APP), and tissue factor pathway inhibitor (TFPI). Kunitz STI protease inhibitor, the trypsin inhibitor initially studied by Moses Kunitz, was extracted from soybeans.

Threonine proteases are a family of proteolytic enzymes harbouring a threonine (Thr) residue within the active site. The prototype members of this class of enzymes are the catalytic subunits of the proteasome, however the acyltransferases convergently evolved the same active site geometry and mechanism.
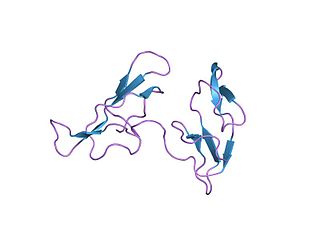
In molecular biology, the Bowman–Birk protease inhibitor family of proteins consists of eukaryotic proteinase inhibitors, belonging to MEROPS inhibitor family I12, clan IF. They mainly inhibit serine peptidases of the S1 family, but also inhibit S3 peptidases.
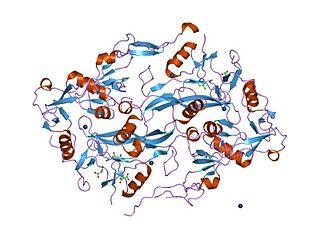
The Kazal domain is an evolutionary conserved protein domain usually indicative of serine protease inhibitors. However, kazal-like domains are also seen in the extracellular part of agrins, which are not known to be protease inhibitors.

In molecular biology, ecotin is a protease inhibitor which belongs to MEROPS inhibitor family I11, clan IN. Ecotins are dimeric periplasmic proteins from Escherichia coli and related Gram-negative bacteria that have been shown to be potent inhibitors of many trypsin-fold serine proteases of widely varying substrate specificity, which belong to MEROPS peptidase family S1. Phylogenetic analysis suggested that ecotin has an exogenous target, possibly neutrophil elastase. Ecotin from E. coli, Yersinia pestis, and Pseudomonas aeruginosa, all species that encounter the mammalian immune system, inhibit neutrophil elastase strongly while ecotin from the plant pathogen Pantoea citrea inhibits neutrophil elastase 1000-fold less potently. Ecotins all potently inhibit pancreatic digestive peptidases trypsin and chymotrypsin, while showing more variable inhibition of the blood peptidases Factor Xa, thrombin, and urokinase-type plasminogen activator.

Protease, serine, 3 is a protein that in humans is encoded by the PRSS3 gene.

The PA clan is the largest group of proteases with common ancestry as identified by structural homology. Members have a chymotrypsin-like fold and similar proteolysis mechanisms but can have identity of <10%. The clan contains both cysteine and serine proteases. PA clan proteases can be found in plants, animals, fungi, eubacteria, archaea and viruses.