
Serine hydroxymethyltransferase (SHMT) is a pyridoxal phosphate (PLP) (Vitamin B6) dependent enzyme (EC 2.1.2.1) which plays an important role in cellular one-carbon pathways by catalyzing the reversible, simultaneous conversions of L-serine to glycine and tetrahydrofolate (THF) to 5,10-Methylenetetrahydrofolate (5,10-CH2-THF). This reaction provides the largest part of the one-carbon units available to the cell.

Thiolases, also known as acetyl-coenzyme A acetyltransferases (ACAT), are enzymes which convert two units of acetyl-CoA to acetoacetyl CoA in the mevalonate pathway.

5,10-Methylenetetrahydrofolate (N5,N10-Methylenetetrahydrofolate; 5,10-CH2-THF) is cofactor in several biochemical reactions. It exists in nature as the diastereoisomer [6R]-5,10-methylene-THF.

Bifunctional purine biosynthesis protein PURH is a protein that in humans is encoded by the ATIC gene.

10-Formyltetrahydrofolate (10-CHO-THF) is a form of tetrahydrofolate that acts as a donor of formyl groups in anabolism. In these reactions 10-CHO-THF is used as a substrate in formyltransferase reactions.
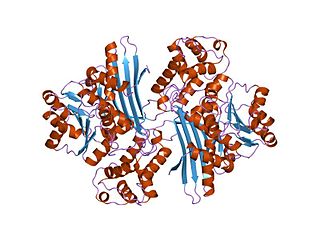
In molecular biology, the protein domain Saccharopine dehydrogenase (SDH), also named Saccharopine reductase, is an enzyme involved in the metabolism of the amino acid lysine, via an intermediate substance called saccharopine. The Saccharopine dehydrogenase enzyme can be classified under EC 1.5.1.7, EC 1.5.1.8, EC 1.5.1.9, and EC 1.5.1.10. It has an important function in lysine metabolism and catalyses a reaction in the alpha-Aminoadipic acid pathway. This pathway is unique to fungal organisms therefore, this molecule could be useful in the search for new antibiotics. This protein family also includes saccharopine dehydrogenase and homospermidine synthase. It is found in prokaryotes, eukaryotes and archaea.
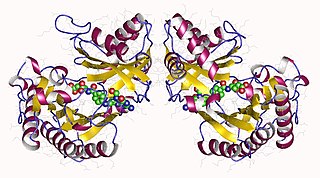
Glutamate formimidoyltransferase is a methyltransferase enzyme which uses tetrahydrofolate as part of histidine catabolism. It catalyses two reactions:
In enzymology, a formimidoyltetrahydrofolate cyclodeaminase (EC 4.3.1.4) is an enzyme that catalyzes the chemical reaction

The enzyme Acid-Induced Arginine Decarboxylase (AdiA), also commonly referred to as arginine decarboxylase, catalyzes the conversion of L-arginine into agmatine and carbon dioxide. The process consumes a proton in the decarboxylation and employs a pyridoxal-5'-phosphate (PLP) cofactor, similar to other enzymes involved in amino acid metabolism, such as ornithine decarboxylase and glutamine decarboxylase. It is found in bacteria and virus, though most research has so far focused on forms of the enzyme in bacteria. During the AdiA catalyzed decarboxylation of arginine, the necessary proton is consumed from the cell cytoplasm which helps to prevent the over-accumulation of protons inside the cell and serves to increase the intracellular pH. Arginine decarboxylase is part of an enzymatic system in Escherichia coli, Salmonella Typhimurium, and methane-producing bacteria Methanococcus jannaschii that makes these organisms acid resistant and allows them to survive under highly acidic medium.

In enzymology, a formate—tetrahydrofolate ligase is an enzyme that catalyzes the chemical reaction
Tyrosine—tRNA ligase, also known as tyrosyl-tRNA synthetase is an enzyme that is encoded by the gene YARS. Tyrosine—tRNA ligase catalyzes the chemical reaction
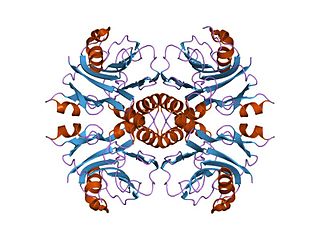
In enzymology, an IMP cyclohydrolase (EC 3.5.4.10) is an enzyme that catalyzes the chemical reaction

In enzymology, a methenyltetrahydrofolate cyclohydrolase (EC 3.5.4.9) is an enzyme that catalyzes the chemical reaction

In enzymology, a formylmethanofuran-tetrahydromethanopterin N-formyltransferase is an enzyme that catalyzes the chemical reaction

MTHFD1 is a gene located in humans on chromosome 14 that encodes for a protein with three distinct enzymatic activities. C-1-tetrahydrofolate synthase, cytoplasmic also known as C1-THF synthase is an enzyme that in humans is encoded by the MTHFD1 gene.

Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial is an enzyme that in humans is encoded by the MTHFD2 gene.

5,10-Methenyltetrahydrofolate (5,10-CH=THF) is a form of tetrahydrofolate that is an intermediate in metabolism. 5,10-CH=THF is a coenzyme that accepts and donates methenyl (CH=) groups.

Phosphoribosylglycinamide formyltransferase (EC 2.1.2.2, 2-amino-N-ribosylacetamide 5'-phosphate transformylase, GAR formyltransferase, GAR transformylase, glycinamide ribonucleotide transformylase, GAR TFase, 5,10-methenyltetrahydrofolate:2-amino-N-ribosylacetamide ribonucleotide transformylase) is an enzyme with systematic name 10-formyltetrahydrofolate:5'-phosphoribosylglycinamide N-formyltransferase. This enzyme catalyses the following chemical reaction

Monofunctional C1-tetrahydrofolate synthase, mitochondrial also known as formyltetrahydrofolate synthetase, is an enzyme that in humans is encoded by the MTHFD1L gene.
UDP-4-amino-4-deoxy-L-arabinose formyltransferase is an enzyme with systematic name 10-formyltetrahydrofolate:UDP-4-amino-4-deoxy-beta-L-arabinose N-formyltransferase. This enzyme catalyses the following chemical reaction

















